
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,100,260 – 5,100,468 |
| Length | 208 |
| Max. P | 0.906754 |

| Location | 5,100,260 – 5,100,353 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -16.16 |
| Energy contribution | -15.83 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5100260 93 + 23771897 -------------GCACACAUAUACAAUGUUACCAAUACCUAGCCAAUAUGGCUGCAAUAGCGCCCUAACCUCCUCCGCCAUAUUCCAACC-CUUUUUUGGGGGGUU -------------....((((.....))))...........(((((((((((((((....)))..............))))))))....((-((.....)))))))) ( -17.13) >DroSec_CAF1 293333 105 + 1 CACACACACAAAUACACACAUGUACAAUGUUACCAAUACCUAGCCAAUAUGGCUGAAAUAACCCCCUAGC--CCUCCGCCAUUUUCUAGCCACCUUUUUGGGUGGUU .................((((.....))))..........(((((.....)))))...............--...............((((((((....)))))))) ( -18.50) >DroSim_CAF1 314944 99 + 1 ------CACAAAUACACACAUGUACAAUGUUACCAAUACCUAGCCAAUAUGGCUGAAAUAGCCCCCUAGC--CCUCCGCCAUUUUCUAGCCCCCUUUUUUGGGGGUU ------...........((((.....)))).........((((.....(((((.((....((......))--..)).)))))...))))(((((......))))).. ( -21.80) >consensus ______CACAAAUACACACAUGUACAAUGUUACCAAUACCUAGCCAAUAUGGCUGAAAUAGCCCCCUAGC__CCUCCGCCAUUUUCUAGCC_CCUUUUUGGGGGGUU .................((((.....))))..........(((((.....)))))................................(((((((......))))))) (-16.16 = -15.83 + -0.33)


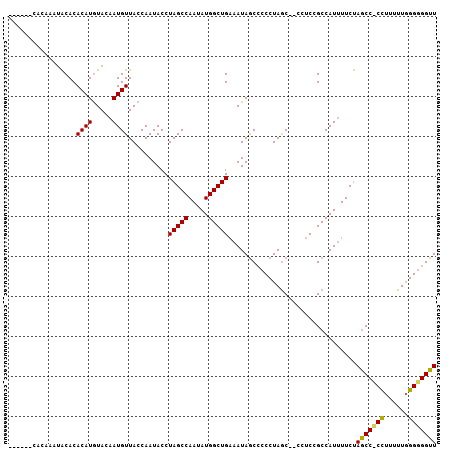
| Location | 5,100,275 – 5,100,393 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -19.59 |
| Energy contribution | -21.15 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5100275 118 - 23771897 UUGCGGUUAAAUGGGCAGUGAGGGGAGGGGGGAGCACCCAAACCCCCCAAAAAAG-GGUU-GGAAUAUGGCGGAGGAGGUUAGGGCGCUAUUGCAGCCAUAUUGGCUAGGUAUUGGUAAC ((((.(.((..(((.(((((.((((.((.(((....)))...)))))).......-((((-(.(..((((((.............))))))).))))).))))).)))..)).).)))). ( -39.22) >DroSec_CAF1 293361 111 - 1 UUGCGGUUAAAUGGGCAGUA------UGGGGGAGCUCCCAAACCACCCAAAAAGGUGGCU-AGAAAAUGGCGGAGG--GCUAGGGGGUUAUUUCAGCCAUAUUGGCUAGGUAUUGGUAAC ((((.(.((..(((.(((((------(((.(((((((((...(((((......)))))..-......((((.....--))))))))))...)))..)))))))).)))..)).).)))). ( -40.10) >DroSim_CAF1 314966 110 - 1 UUGCGGUUAAAUGGGCAGUG-------GGAGGAAAACCCAAACCCCCAAAAAAGGGGGCU-AGAAAAUGGCGGAGG--GCUAGGGGGCUAUUUCAGCCAUAUUGGCUAGGUAUUGGUAAC ((((.(.((..(((.(((((-------((..((((.(((...(((((......)))))..-......((((.....--)))).)))....))))..)).))))).)))..)).).)))). ( -35.60) >DroYak_CAF1 305645 101 - 1 UUGCGGUUAAAUGGGCAGUG------UGGGGAAGCU----------CCAAAAGGGGGGCUCGGAAAAUGGCGGAGG--GCUAGGG-GAUAUUUCAGCCAUAUUGGCUAGGUAUUGGUAAC ((((.(.((..(((.(((((------(((.((((((----------((......)))))........((((.....--))))...-.....)))..)))))))).)))..)).).)))). ( -31.00) >consensus UUGCGGUUAAAUGGGCAGUG______UGGGGGAGCACCCAAACCCCCCAAAAAGGGGGCU_AGAAAAUGGCGGAGG__GCUAGGGGGCUAUUUCAGCCAUAUUGGCUAGGUAUUGGUAAC ((((.(.((..(((.((((.......................(((((......)))))........(((((.((((..((......))..)))).))))))))).)))..)).).)))). (-19.59 = -21.15 + 1.56)


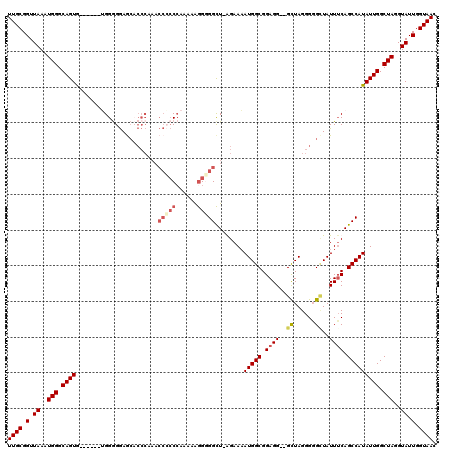
| Location | 5,100,353 – 5,100,468 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -25.83 |
| Energy contribution | -25.61 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5100353 115 + 23771897 UGGGUGCUCCCCCCUCCCCUCACUGCCCAUUUAACCGCAAGCCAUGCUUCUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUGCUUUUCCUCUCUCGC (((((...................)))))...(((((((.((..(((((...(((((.....))))).......)))))...)))))))))........................ ( -26.71) >DroSec_CAF1 293438 109 + 1 UGGGAGCUCCCCCA------UACUGCCCAUUUAACCGCAAGCCAUGCUUCUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUGCUUUUCCUCUCGCGC ((((......))))------....((......(((((((.((..(((((...(((((.....))))).......)))))...)))))))))........)).............. ( -27.74) >DroSim_CAF1 315043 108 + 1 UGGGUUUUCCUCC-------CACUGCCCAUUUAACCGCAAGCCAUGCUUCUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUGCUUUUCCUCUCUCGC (((((........-------....)))))...(((((((.((..(((((...(((((.....))))).......)))))...)))))))))........................ ( -27.30) >consensus UGGGUGCUCCCCC_______CACUGCCCAUUUAACCGCAAGCCAUGCUUCUAAUGCCGCGUUGGCAUAAAUUUAAAGCAGCAGCUGCGGUUUUUCCACUGCUUUUCCUCUCUCGC .(((....))).............((......(((((((.((..(((((...(((((.....))))).......)))))...)))))))))........)).............. (-25.83 = -25.61 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:25 2006