
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,099,354 – 5,099,468 |
| Length | 114 |
| Max. P | 0.625305 |

| Location | 5,099,354 – 5,099,468 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.06 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5099354 114 + 23771897 CUAGAUGGCGCCAGCACGUGCUCCGAGUCGGGGGAUUCCCCUCUCUAUUUUCCACCCCC------CUCUACUCUACGCACCCAAUCAAUACUUGAAUUGGUUUCGGUUUACCAAACUUUU .....(((.(((.....((((...((((.(((((.....................))))------)...))))...))))(((((((.....)).)))))....)))...)))....... ( -27.00) >DroSec_CAF1 292423 119 + 1 CUAGAUGGCGUCAGCACGUGCUCCGGGUCGGGGGAUUCCCCUCUCUUUUUUUCACCCCCCUCCCCCUCUACUC-ACGCACCCAAUCAAUAUUUGAAUUGGUUUCGGUUUACCAACCUUUU .......((((.(((....)))..(((..(((((.....................)))))..)))........-))))..(((((((.....)).)))))....((((....)))).... ( -27.60) >DroSim_CAF1 314067 118 + 1 CUAGAUGGCGUCAGCACGUGCUCCCAGUCGGGGGAUUCCCC-CUCUUUUUUCCACCCCCCUCCCCCUCUACUC-ACGCACCCAAUCAAUAUUUGAAUUGGUUUCGGUUUACCAACCUUUU .((((.((((..(((....)))..).)))((((((......-..................))))))))))...-......(((((((.....)).)))))....((((....)))).... ( -22.66) >DroYak_CAF1 304711 115 + 1 CUAGAUGGCGCCAGCACGUGCUCCCUGUCAGGGGAUUCCCC-CUCUUUUCUACACCCCC---UCCCUCUAUUC-GUGAAACCAAUCAAUAUUUGAAUUGGUUUCGGUUUACCAACCUUUU .(((((((((..(((....)))...)))))((((....)))-).....)))).......---...........-..(((((((((((.....)).)))))))))((((....)))).... ( -28.20) >consensus CUAGAUGGCGCCAGCACGUGCUCCCAGUCGGGGGAUUCCCC_CUCUUUUUUCCACCCCC___CCCCUCUACUC_ACGCACCCAAUCAAUAUUUGAAUUGGUUUCGGUUUACCAACCUUUU .((((.(((...(((....)))....)))(((((....................))))).......))))..........(((((((.....)).)))))....((....))........ (-17.90 = -18.15 + 0.25)

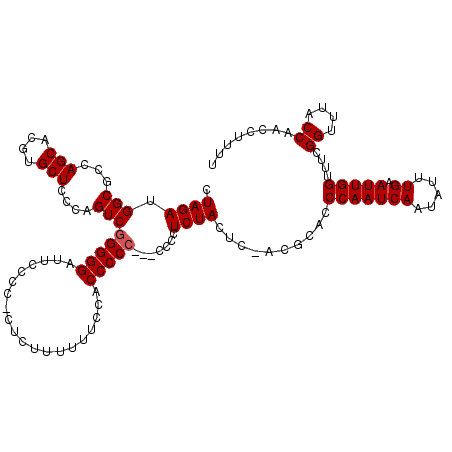
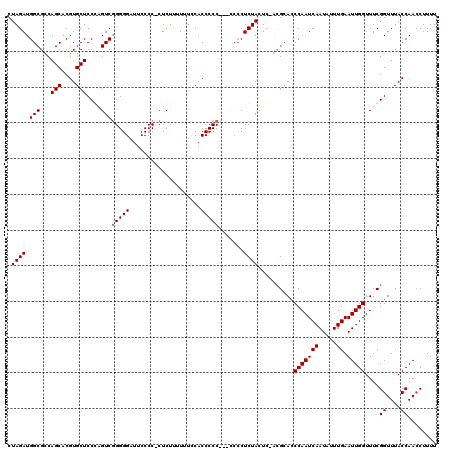
| Location | 5,099,354 – 5,099,468 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.06 |
| Mean single sequence MFE | -36.51 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.07 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5099354 114 - 23771897 AAAAGUUUGGUAAACCGAAACCAAUUCAAGUAUUGAUUGGGUGCGUAGAGUAGAG------GGGGGUGGAAAAUAGAGAGGGGAAUCCCCCGACUCGGAGCACGUGCUGGCGCCAUCUAG ...((..((((...(((...((((((........))))))((((...((((...(------((((((.................))))))).))))...))))....))).)))).)).. ( -36.03) >DroSec_CAF1 292423 119 - 1 AAAAGGUUGGUAAACCGAAACCAAUUCAAAUAUUGAUUGGGUGCGU-GAGUAGAGGGGGAGGGGGGUGAAAAAAAGAGAGGGGAAUCCCCCGACCCGGAGCACGUGCUGACGCCAUCUAG ...((((.(((.........((((((........))))))(..(((-(..(....(((..(((((((.................)))))))..))).)..))))..)....))))))).. ( -39.53) >DroSim_CAF1 314067 118 - 1 AAAAGGUUGGUAAACCGAAACCAAUUCAAAUAUUGAUUGGGUGCGU-GAGUAGAGGGGGAGGGGGGUGGAAAAAAGAG-GGGGAAUCCCCCGACUGGGAGCACGUGCUGACGCCAUCUAG ...((((.(((...((....((((((........)))))).(((..-..)))..))((..(((((((...........-.....)))))))..))(..(((....)))..)))))))).. ( -34.79) >DroYak_CAF1 304711 115 - 1 AAAAGGUUGGUAAACCGAAACCAAUUCAAAUAUUGAUUGGUUUCAC-GAAUAGAGGGA---GGGGGUGUAGAAAAGAG-GGGGAAUCCCCUGACAGGGAGCACGUGCUGGCGCCAUCUAG ....((((....))))((((((((((........))))))))))..-........(((---...(((((.......((-(((....))))).......(((....))).))))).))).. ( -35.70) >consensus AAAAGGUUGGUAAACCGAAACCAAUUCAAAUAUUGAUUGGGUGCGU_GAGUAGAGGGG___GGGGGUGGAAAAAAGAG_GGGGAAUCCCCCGACUCGGAGCACGUGCUGACGCCAUCUAG ......((((....))))..((((((........)))))).....................(((((((...........((((....)))).......(((....)))..)))).))).. (-23.26 = -23.07 + -0.19)

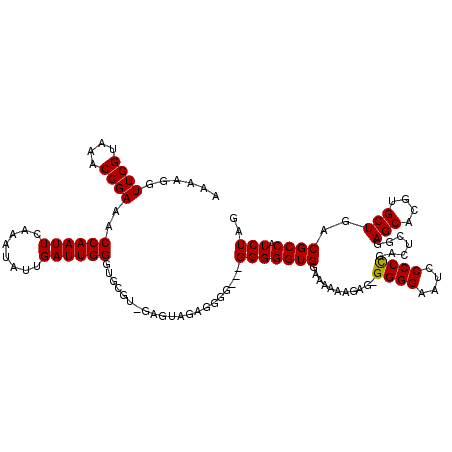
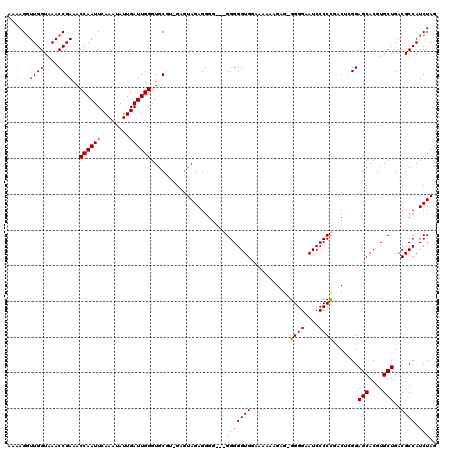
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:22 2006