
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,098,794 – 5,098,947 |
| Length | 153 |
| Max. P | 0.995708 |

| Location | 5,098,794 – 5,098,908 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.99 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5098794 114 + 23771897 CAAGAUGAACAA------GUUUGUCUUGCAGCAAAGAUGUUUACCGACUGCCAAUGCAAAAAAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACAAAAUGGCAAACGUUUCCCAG (((((..((...------.))..)))))......((((((((.(((...(((........................)))((((((....))))))........))).))))))))..... ( -25.86) >DroSec_CAF1 291867 113 + 1 CAAGAUGUACAA------GUUUGUCUUGCAGCAAAGAUGUUUACCGACUGCCAAUGCAAA-AAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACAAAAUGGCAAACGUUUUCCAG (((((((.....------...))))))).....(((((((((.(((...(((........-...............)))((((((....))))))........))).))))))))).... ( -24.10) >DroSim_CAF1 313519 104 + 1 ---------CAA------GUUUGUCUUGCAGCAAAGAUGUUUACCGACUGCCAAUGCAAA-AAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACGAAAUGGCAAACGUUUUCCAG ---------...------(((((((..((.....(((((.........(((....)))..-......))))).....))((((((....)))))).........)))))))......... ( -21.83) >DroYak_CAF1 304128 118 + 1 CUAGAUGUACAAGUACGAGUAUGUCUUGCAGCAAAGAUGUUUACCGACUGCCAAUGCAAA-AAAUAUCAUUUAAAUGGCAUCACCAAAUGGAGAUACACA-AAUGGCAAACGUUUUCCAG ..((((((((........)))))))).......(((((((((.(((...((((.......-..............))))(((.((....)).))).....-..))).))))))))).... ( -21.90) >consensus CAAGAUGUACAA______GUUUGUCUUGCAGCAAAGAUGUUUACCGACUGCCAAUGCAAA_AAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACAAAAUGGCAAACGUUUUCCAG ..................(((((((..((...((((((((((......(((....)))...))))))).))).....))((((((....)))))).........)))))))......... (-20.30 = -20.80 + 0.50)

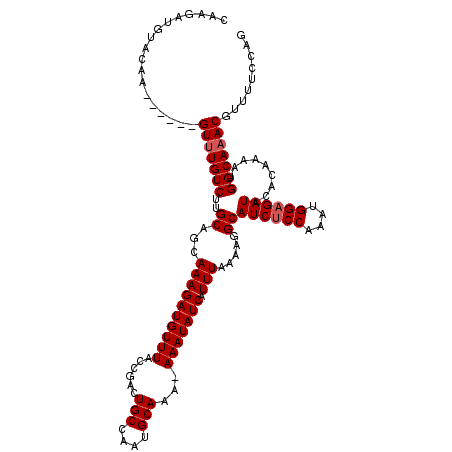
| Location | 5,098,794 – 5,098,908 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.99 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -23.33 |
| Energy contribution | -23.57 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5098794 114 - 23771897 CUGGGAAACGUUUGCCAUUUUGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUUUUUUGCAUUGGCAGUCGGUAAACAUCUUUGCUGCAAGACAAAC------UUGUUCAUCUUG ...(....)(((((((.......((((((((....))))))))..................((((....))))..))))))).....((..(((((.....)------)))).))..... ( -29.10) >DroSec_CAF1 291867 113 - 1 CUGGAAAACGUUUGCCAUUUUGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUU-UUUGCAUUGGCAGUCGGUAAACAUCUUUGCUGCAAGACAAAC------UUGUACAUCUUG ..(((....(((((((.......((((((((....))))))))................-.((((....))))..))))))).....((.((((((.....)------)))))))))).. ( -29.40) >DroSim_CAF1 313519 104 - 1 CUGGAAAACGUUUGCCAUUUCGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUU-UUUGCAUUGGCAGUCGGUAAACAUCUUUGCUGCAAGACAAAC------UUG--------- .........(((((.(((((...((((((((....)))))))).....))))).....(-((((((...((((..(((....))).))))))))))))))))------...--------- ( -25.40) >DroYak_CAF1 304128 118 - 1 CUGGAAAACGUUUGCCAUU-UGUGUAUCUCCAUUUGGUGAUGCCAUUUAAAUGAUAUUU-UUUGCAUUGGCAGUCGGUAAACAUCUUUGCUGCAAGACAUACUCGUACUUGUACAUCUAG (((((....(((((((...-((.(((((.((....)).)))))))..............-.((((....))))..))))))).....((.((((((((......)).))))))))))))) ( -31.00) >consensus CUGGAAAACGUUUGCCAUUUUGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUU_UUUGCAUUGGCAGUCGGUAAACAUCUUUGCUGCAAGACAAAC______UUGUACAUCUUG .........(((((((.....(.((((((((....))))))))).................((((....))))..))))))).((((......))))....................... (-23.33 = -23.57 + 0.25)



| Location | 5,098,828 – 5,098,947 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -24.31 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5098828 119 + 23771897 UUACCGACUGCCAAUGCAAAAAAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACAAAAUGGCAAACGUUUCCCAGGGGCAGUCAAGCGAUGUAAAAUAUUUGUA-UGGAGAGGUU ..((((((((((..((..(((.....((((((.....(.((((((....)))))).)...))))))......)))..))..)))))))..((((..........)))).-......))). ( -29.70) >DroSec_CAF1 291901 118 + 1 UUACCGACUGCCAAUGCAAA-AAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACAAAAUGGCAAACGUUUUCCAGGGGCAGUCAAGCGAUGUAAAACAUAUGUA-CGGAGAGGUU ..((((((((((..((.(((-(....((((((.....(.((((((....)))))).)...))))))......)))).))..)))))))..((((((.....))).))).-......))). ( -31.60) >DroSim_CAF1 313544 118 + 1 UUACCGACUGCCAAUGCAAA-AAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACGAAAUGGCAAACGUUUUCCAGCGGCAGUCAAGCGAUGUAAAAUAUAUGUA-CGGGGAGGUU ..((((((((((..((.(((-(....((((((.....(.((((((....)))))).)...))))))......)))).))..)))))))...(((((((.....))))).-))....))). ( -31.00) >DroYak_CAF1 304168 118 + 1 UUACCGACUGCCAAUGCAAA-AAAUAUCAUUUAAAUGGCAUCACCAAAUGGAGAUACACA-AAUGGCAAACGUUUUCCAGUGGCAGUCAAGCGAUGUAAAAUAUGUGGAGUGGACAGAGG ...((((((((((.((.(((-(....((((((...((..(((.((....)).))).)).)-)))))......)))).)).))))))))...(.(((((...))))).)...))....... ( -25.40) >consensus UUACCGACUGCCAAUGCAAA_AAAUAUCAUUUAAAAGGCAUCUCCAAAUGGAGAUACACAAAAUGGCAAACGUUUUCCAGGGGCAGUCAAGCGAUGUAAAAUAUAUGUA_CGGAGAGGUU ...(((((((((..((.(((......((((((.....(.((((((....)))))).)...)))))).......))).))..)))))))..(((((((...)))).)))...))....... (-24.31 = -25.12 + 0.81)



| Location | 5,098,828 – 5,098,947 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -29.58 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5098828 119 - 23771897 AACCUCUCCA-UACAAAUAUUUUACAUCGCUUGACUGCCCCUGGGAAACGUUUGCCAUUUUGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUUUUUUGCAUUGGCAGUCGGUAA ..........-.................(((.(((((((..((.((((....((.(((((.(.((((((((....)))))))))....))))).))...)))).))..)))))))))).. ( -32.00) >DroSec_CAF1 291901 118 - 1 AACCUCUCCG-UACAUAUGUUUUACAUCGCUUGACUGCCCCUGGAAAACGUUUGCCAUUUUGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUU-UUUGCAUUGGCAGUCGGUAA .(((.....(-((.........))).......(((((((..((.((((.((....(((((.(.((((((((....)))))))))....)))))..)).)-))).))..)))))))))).. ( -33.00) >DroSim_CAF1 313544 118 - 1 AACCUCCCCG-UACAUAUAUUUUACAUCGCUUGACUGCCGCUGGAAAACGUUUGCCAUUUCGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUU-UUUGCAUUGGCAGUCGGUAA .(((.....(-((.........))).......((((((((.((.((((.((....(((((...((((((((....)))))))).....)))))..)).)-))).)).))))))))))).. ( -33.60) >DroYak_CAF1 304168 118 - 1 CCUCUGUCCACUCCACAUAUUUUACAUCGCUUGACUGCCACUGGAAAACGUUUGCCAUU-UGUGUAUCUCCAUUUGGUGAUGCCAUUUAAAUGAUAUUU-UUUGCAUUGGCAGUCGGUAA ............................(((.((((((((.((.((((.((....((((-((.(((((.((....)).)))))....))))))..)).)-))).)).))))))))))).. ( -30.60) >consensus AACCUCUCCA_UACAUAUAUUUUACAUCGCUUGACUGCCCCUGGAAAACGUUUGCCAUUUUGUGUAUCUCCAUUUGGAGAUGCCUUUUAAAUGAUAUUU_UUUGCAUUGGCAGUCGGUAA ............................(((.((((((((.((.(((.....((.(((((...((((((((....)))))))).....))))).))....))).)).))))))))))).. (-29.58 = -30.32 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:20 2006