
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,083,444 – 5,083,650 |
| Length | 206 |
| Max. P | 0.965228 |

| Location | 5,083,444 – 5,083,556 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5083444 112 - 23771897 --------CCCACAGUUACUUAACAAUAUUUCUCAUUGUUGUUGAGACGUUGCAGGUGUUUGCUGAAGCAUAUCAAAAAUAUAUGUAUGUACGAGCAUAGCUGCGACUACGUCGACGAUU --------......(((.((((((((((........))))))))))..((((((((((((((.((..((((((.......))))))...)))))))))..)))))))......))).... ( -30.30) >DroSec_CAF1 289666 110 - 1 --------CC--CAGUUACUUAACAAUGUUUCUCAUUGUUGUUGAGACGUUGCAGGUGUUUGCUGAAGCAUAUCAAAAAUAUAUGUAUGUACGACCAUAGCUGCGACUACGUCGACGAUU --------..--..(((.((((((((((........))))))))))..(((((((.((((((.((..((((((.......))))))...)))))..))).)))))))......))).... ( -25.30) >DroSim_CAF1 310966 110 - 1 --------CC--CAGUUACUUAACAAUAUUUCUCAUUGUUGUUGAGACGUUGCAGGUGUUUGCUGAAGCAUAUCAAAAAUAUAUGUAUAUACGACCAUAGCUGCGACUACGUCGACGAUU --------..--..(((.((((((((((........))))))))))..(((((((.((((((.....((((((.......)))))).....)))..))).)))))))......))).... ( -26.60) >DroYak_CAF1 301186 118 - 1 CUCCCCUCAC--CAGUUACUUAACAAUAUUUCCCAUUGUUGUUGAGACGUUGCAGGUGUUUGCUGAAGCAUAUCAAAAAUAUAUGUAUAUACAAACAUAGCUGCGACUACGUCGACGACU ..........--..(((.((((((((((........))))))))))..((((((((((((((.....((((((.......)))))).....)))))))..)))))))......))).... ( -30.60) >consensus ________CC__CAGUUACUUAACAAUAUUUCUCAUUGUUGUUGAGACGUUGCAGGUGUUUGCUGAAGCAUAUCAAAAAUAUAUGUAUAUACGACCAUAGCUGCGACUACGUCGACGAUU ..............(((.((((((((((........))))))))))..((((((((((((((.....((((((.......)))))).....)))))))..)))))))......))).... (-28.44 = -28.50 + 0.06)


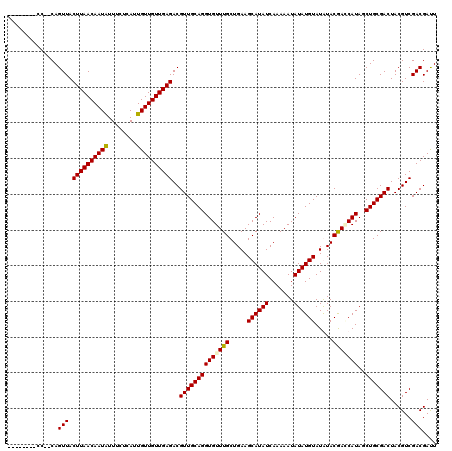
| Location | 5,083,524 – 5,083,623 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5083524 99 + 23771897 AACAAUGAGAAAUAUUGUUAAGUAACUGUGGGGGGGGGGGGGGCUGAGAAUAGUGGAUGCAGUUUUUGGUGCAUUAGCGGAGUGCGUUACCCCCGGCUC (((((((.....)))))))........((.(((((.....(.((..(((((.((....)).)))))..)).)..(((((.....)))))))))).)).. ( -29.90) >DroSec_CAF1 289746 96 + 1 AACAAUGAGAAACAUUGUUAAGUAACUG--GG-GGAGGGGGUACUGAGAGUAGUGGAUGCAGUUUUUGGUGCAUUAGCGGAGUGCGUUACCCCCGGCUC (((((((.....)))))))......(((--((-((.....((((..((((..((....))..))))..))))..(((((.....))))))))))))... ( -32.80) >DroSim_CAF1 311046 97 + 1 AACAAUGAGAAAUAUUGUUAAGUAACUG--GGGGGAGGGGGUACUGAGAGUAGUGGAUGCAGUUUUUGGUGCAUUAGCGGAGUGCGUUACCCCCGGCUC (((((((.....)))))))......(((--((((......((((..((((..((....))..))))..))))..(((((.....))))))))))))... ( -30.60) >consensus AACAAUGAGAAAUAUUGUUAAGUAACUG__GGGGGAGGGGGUACUGAGAGUAGUGGAUGCAGUUUUUGGUGCAUUAGCGGAGUGCGUUACCCCCGGCUC (((((((.....))))))).................(((((((.((......((.((((((.(....).)))))).))......)).)))))))..... (-26.47 = -26.37 + -0.11)


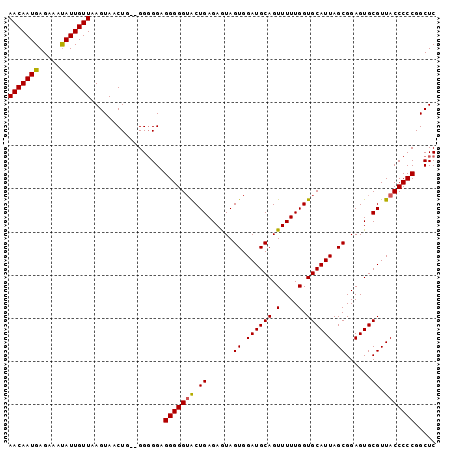
| Location | 5,083,524 – 5,083,623 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -17.80 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5083524 99 - 23771897 GAGCCGGGGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUAUUCUCAGCCCCCCCCCCCCCCACAGUUACUUAACAAUAUUUCUCAUUGUU (((..((((((...((.....))..(((((........)))))............))))))............(((....))).......)))...... ( -18.70) >DroSec_CAF1 289746 96 - 1 GAGCCGGGGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUACUCUCAGUACCCCCUCC-CC--CAGUUACUUAACAAUGUUUCUCAUUGUU .....(((((((..((.....))..(((((........))))).............)))))))...-..--.........(((((((.....))))))) ( -23.00) >DroSim_CAF1 311046 97 - 1 GAGCCGGGGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUACUCUCAGUACCCCCUCCCCC--CAGUUACUUAACAAUAUUUCUCAUUGUU (((..(((((((..((.....))..(((((........))))).............)))))))......--..(((....))).......)))...... ( -20.10) >consensus GAGCCGGGGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUACUCUCAGUACCCCCUCCCCC__CAGUUACUUAACAAUAUUUCUCAUUGUU (((..(((((((..((.....))..(((((........))))).............)))))))..........(((....))).......)))...... (-17.80 = -18.47 + 0.67)


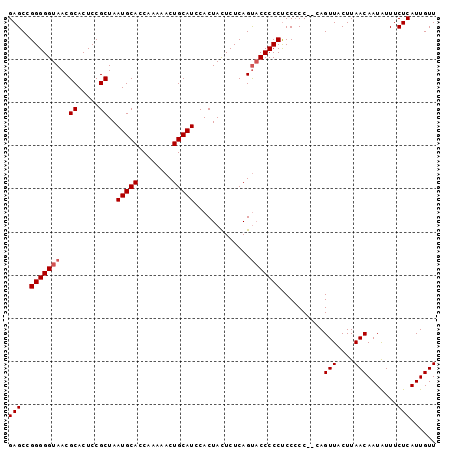
| Location | 5,083,556 – 5,083,650 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5083556 94 - 23771897 ACCCAAAAC-------------GAGGAGCAGAUGGAGCAGGAGCCGG-GGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUAUUCUCAGCCCCCC------CCCCC------ .........-------------..(((((((.(((.(((..(((.((-((........)))))))..))).)))....)))).))).................------.....------ ( -27.30) >DroSec_CAF1 289776 93 - 1 ACCCAAAAC-------------GAGGAGCAGAUGGAGCAGGAGCCGG-GGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUACUCUCAGUACCCC------CUCC------- .........-------------((((.((((.(((.(((..(((.((-((........)))))))..))).)))....))))....(((......)))....)------))).------- ( -28.70) >DroSim_CAF1 311076 94 - 1 ACCCACAAC-------------GAGGAGCAGAUGGAGCAGGAGCCGG-GGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUACUCUCAGUACCCC------CUCCC------ .........-------------((((.((((.(((.(((..(((.((-((........)))))))..))).)))....))))....(((......)))....)------)))..------ ( -28.70) >DroYak_CAF1 301304 120 - 1 ACCCAAAACGAGGACCCCAAACGAGGAGCAGAUGCAGCAGGAGCUGGAGGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUACUCUCAGACCACCCCUCCCCUCCCCCUCCA .........((((...((......)).((....))....((((..((((((....((.....))..(((((........)))))..................)))))).)))).)))).. ( -28.90) >consensus ACCCAAAAC_____________GAGGAGCAGAUGGAGCAGGAGCCGG_GGGUAACGCACUCCGCUAAUGCACCAAAAACUGCAUCCACUACUCUCAGUACCCC______CUCCC______ ........................(((((((.(((.(((..(((.((.((........)))))))..))).)))....)))).))).................................. (-22.25 = -22.50 + 0.25)

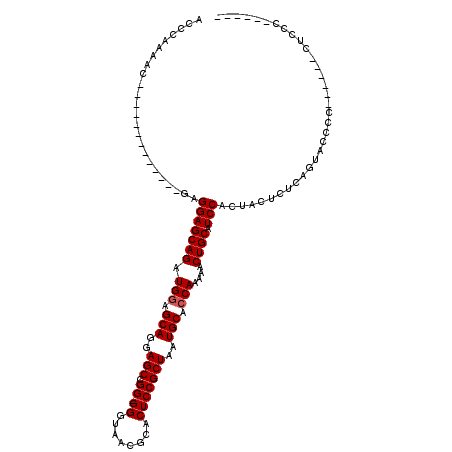
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:14 2006