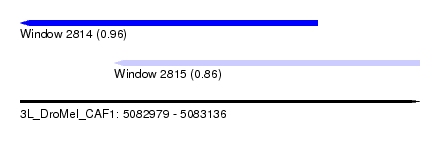
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,082,979 – 5,083,136 |
| Length | 157 |
| Max. P | 0.964004 |

| Location | 5,082,979 – 5,083,096 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
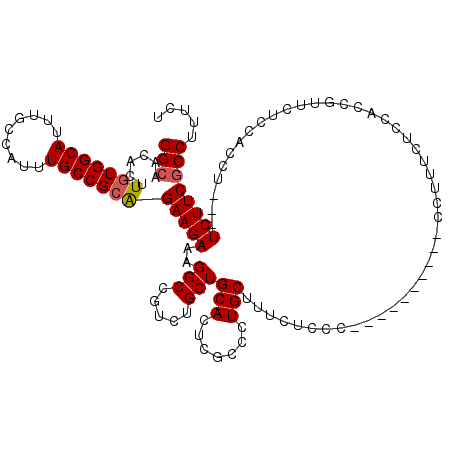
>3L_DroMel_CAF1 5082979 117 - 23771897 GGCAACACUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCUCCUUUCUCCUCCUUUCUCCACCGUUCUCCACCU---UCUUCGCCUUUCU (((.....((((((((.........)))))))).(((((((......((.((....))...))........)))))))..........................---.....)))..... ( -24.94) >DroSec_CAF1 289258 107 - 1 GGCAACUCUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC----------CAUUUCCCCACCGUUCUGCACCU---UCUUCGCCUUUCU (((((.....(((((....)))))))))).....((((((((.....(.((((..((..............----------...........))...)))).).---.....)))))))) ( -25.11) >DroSim_CAF1 310604 107 - 1 GGCAACACUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC----------CCUUUCUCCACCGUUCUCCACCU---UCUUCGCCUUUCU (((......(((((((.........)))))))(.(((((((......((.((....))...))........----------))))))).)..............---.....)))..... ( -25.34) >DroYak_CAF1 300738 110 - 1 GGCAACUCUUUUGGCAUUUAACAUUUGCCGUGGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC----------CCUUUCUCCACAUUACUCCACCUUUCUCUUCACCUCUCU ((..........((((.........))))(((((.((((((......((.((....))...))........----------)))))))))))......)).................... ( -23.54) >consensus GGCAACACUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC__________CCUUUCUCCACCGUUCUCCACCU___UCUUCGCCUUUCU (((......(((((((.........)))))))(((((..(((.....)))(((.......)))............................................))))))))..... (-19.94 = -20.25 + 0.31)


| Location | 5,083,016 – 5,083,136 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -27.85 |
| Energy contribution | -28.47 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5083016 120 - 23771897 UCUAGUUCCAAGUUUGCGAACCCAUUUUAGCCCGAAAGUAGGCAACACUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCUCCUUUCUCC ....((((((....)).))))............((((((((((.......(((((....))))).(((.(((((.(......).))))).)))...).)))))))))............. ( -33.00) >DroSec_CAF1 289294 109 - 1 UCUAGUUCCAA--UUGCGAACUCAUUUCAGCCCGAAAGUAGGCAACUCUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC--------- ...((((((..--..).)))))...........((((((((((.......(((((....))))).(((.(((((.(......).))))).)))...).)))))))))....--------- ( -33.30) >DroSim_CAF1 310640 109 - 1 UCUAGUUCCAA--UUGCGAACUCAUUUUAGCCCGAAAGUAGGCAACACUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC--------- ...((((((..--..).)))))...........((((((((((.......(((((....))))).(((.(((((.(......).))))).)))...).)))))))))....--------- ( -33.30) >DroYak_CAF1 300777 109 - 1 ACUAGUUCCUA--UUGCGAACUCAUUUCAGCCCAAAAGUAGGCAACUCUUUUGGCAUUUAACAUUUGCCGUGGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC--------- ...........--..((((..........(((.(((((.((....))))))))))..........(((.(..((.(......).))..).))).)))).............--------- ( -23.60) >consensus UCUAGUUCCAA__UUGCGAACUCAUUUCAGCCCGAAAGUAGGCAACACUUGUGGCAUUUGCCAUUUGCCGCAGAAGAAAGGCCGUCUGCUGCACUCGCCCUGCUUUCUCCC_________ ....(((((......).))))............((((((((((.......(((((....))))).(((.(((((.(......).))))).)))...).)))))))))............. (-27.85 = -28.47 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:10 2006