
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,081,555 – 5,081,865 |
| Length | 310 |
| Max. P | 0.992241 |

| Location | 5,081,555 – 5,081,675 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -28.49 |
| Energy contribution | -29.49 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5081555 120 - 23771897 CCGGAAAUGCAACAGGAAAUCAGUGUGCAAGGGCAGUAAAUGCUGAUGCCACGCCCACAAAACGUCCAGCGAACAACGCCCACCACACCCCGCCACAUGCGGCACGUUUCCACAUUGCAC .......(((((..((((((..(((((...((((.((...((((((((..............))).)))))....))))))..))))).((((.....))))...))))))...))))). ( -33.64) >DroSec_CAF1 287820 120 - 1 CCGGAAAUGCAACAGGAAAUCAGUGUGCAAGGGCAGUAAAUGCUGAUGCCACGCCCACAAAACGUCCAGCGAACAACGCCCAUCACACCCCGCCACAUGCGGCACGUUUCCACAUUGCAC .......(((((..((((((..(((((...((((.((...((((((((..............))).)))))....))))))..))))).((((.....))))...))))))...))))). ( -33.64) >DroSim_CAF1 309164 120 - 1 CCGGAAAUGCAACAGGAAAUCAGUGUGCAAGGGCAGUAAAUGCUGAUGCCACGCCCACAGAACGUCCAGCGAACAACGCCCAUCACACCCCGCCACAUGCGGCACGUUUCCACAUUGCAC .......(((((..((((((..(((((...((((.((...((((((((..............))).)))))....))))))..))))).((((.....))))...))))))...))))). ( -33.64) >DroYak_CAF1 299263 120 - 1 CCGGAAAUGCAACAGGAAAUCAGUGUGCAAGGGCAGUAAAUGCUGUUGCCACACCCACAAAACGUACAGCGAACUACGCCCACCACAACCCACCACAUGCGGCACGUUCCCACAUUGCAC .......(((((..((......((((((((.((((.....)))).))).)))))......(((((...(((.....)))...((.((..........)).)).))))).))...))))). ( -27.90) >consensus CCGGAAAUGCAACAGGAAAUCAGUGUGCAAGGGCAGUAAAUGCUGAUGCCACGCCCACAAAACGUCCAGCGAACAACGCCCACCACACCCCGCCACAUGCGGCACGUUUCCACAUUGCAC .......(((((..((((((..(((((...((((.((...((((((((..............))).)))))....))))))..))))).((((.....))))...))))))...))))). (-28.49 = -29.49 + 1.00)

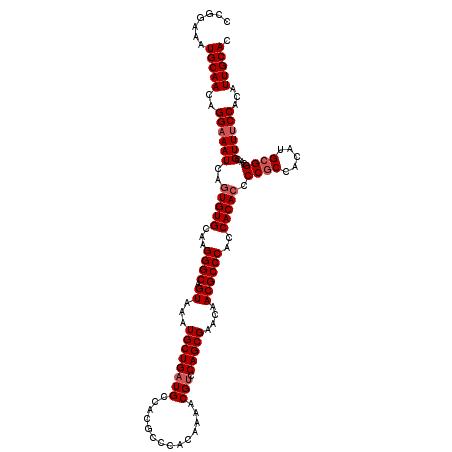
| Location | 5,081,675 – 5,081,785 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -21.11 |
| Consensus MFE | -18.08 |
| Energy contribution | -19.45 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5081675 110 + 23771897 UUAGCCAACGAUGAAGUUGCCUCCUUUCCCACCCGAAUGGCCUUUCCCUUUUCCAUUCGGGCAUCCACA--------CACUUUGUGUU--AGCACAUAAAACUAAAUCGAAUUUAGAACA ........((((..((((.............(((((((((............)))))))))........--------.....((((..--..))))...))))..))))........... ( -23.70) >DroSec_CAF1 287940 110 + 1 UUAGCCAACGAUGAAGUUGCCUCCUCUCCCACCCGAAUGGCCUUUCCCUUUUCCAUUCGGGCAUCCACA--------CACUUUGUGUU--AGCACAUAAAACUAAAUCGAAUUUAGAACA ........((((..((((.............(((((((((............)))))))))........--------.....((((..--..))))...))))..))))........... ( -23.70) >DroSim_CAF1 309284 110 + 1 UUAGCCAACGAUGAAGUUGCCUCCUUUCCCACCCGAAUGGCCUUUCCCUUUUCCAUUCGGGCAUCCACA--------CACUUUGUGUU--AGCACAUAAAACUAAAUCGAAUUUAGAACA ........((((..((((.............(((((((((............)))))))))........--------.....((((..--..))))...))))..))))........... ( -23.70) >DroYak_CAF1 299383 120 + 1 UCAGCCAACGAUGAAGUGGCCUCCUUUCCCACCAGCAAGUCCUUUCCCUUUUCCAUUUGAACUUCCACACACAUACACACUUUGUGCUAUAGCACAUAAAACUAAAUCGAAUUUAGAACA ........((((((((((((..............))((((.(................).)))).............))))))((((....))))..........))))........... ( -13.33) >consensus UUAGCCAACGAUGAAGUUGCCUCCUUUCCCACCCGAAUGGCCUUUCCCUUUUCCAUUCGGGCAUCCACA________CACUUUGUGUU__AGCACAUAAAACUAAAUCGAAUUUAGAACA ........((((..((((.............(((((((((............))))))))).....................(((((....)))))...))))..))))........... (-18.08 = -19.45 + 1.37)


| Location | 5,081,715 – 5,081,825 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5081715 110 + 23771897 CCUUUCCCUUUUCCAUUCGGGCAUCCACA--------CACUUUGUGUU--AGCACAUAAAACUAAAUCGAAUUUAGAACACUUCCAAAGGAAAUUGGAAAUUAACUAAAAAUGAGUUAUU ..(((((..(((((.((.((((...((((--------.....))))..--.))........((((((...)))))).......)).)))))))..))))).(((((.......))))).. ( -19.70) >DroSec_CAF1 287980 110 + 1 CCUUUCCCUUUUCCAUUCGGGCAUCCACA--------CACUUUGUGUU--AGCACAUAAAACUAAAUCGAAUUUAGAACACUUCCAAAGGAAAUUGGAAAUUAACUAAAAAUGAGUUAGU ...((((..(((((.((.((((...((((--------.....))))..--.))........((((((...)))))).......)).)))))))..))))(((((((.......))))))) ( -20.60) >DroSim_CAF1 309324 110 + 1 CCUUUCCCUUUUCCAUUCGGGCAUCCACA--------CACUUUGUGUU--AGCACAUAAAACUAAAUCGAAUUUAGAACACUUCCAAAGGAAAUUGGAAAUUAACUAAAAAUGAGUUAUU ..(((((..(((((.((.((((...((((--------.....))))..--.))........((((((...)))))).......)).)))))))..))))).(((((.......))))).. ( -19.70) >DroYak_CAF1 299423 120 + 1 CCUUUCCCUUUUCCAUUUGAACUUCCACACACAUACACACUUUGUGCUAUAGCACAUAAAACUAAAUCGAAUUUAGAACACUUCCAAAGGAAAUUGGAAAUUAACUAAAAAUGAGUUAUU ..(((((..(((((.((((.......................(((((....))))).....((((((...))))))........)))))))))..))))).(((((.......))))).. ( -22.60) >consensus CCUUUCCCUUUUCCAUUCGGGCAUCCACA________CACUUUGUGUU__AGCACAUAAAACUAAAUCGAAUUUAGAACACUUCCAAAGGAAAUUGGAAAUUAACUAAAAAUGAGUUAUU ..(((((..(((((............................(((((....))))).....((((((...))))))............)))))..))))).(((((.......))))).. (-19.64 = -19.45 + -0.19)



| Location | 5,081,715 – 5,081,825 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -21.95 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5081715 110 - 23771897 AAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU--AACACAAAGUG--------UGUGGAUGCCCGAAUGGAAAAGGGAAAGG ..(((((.......))))).(((((..((((((((((..((((((...........((((.......)))--)((((.....)--------)))))))))))))).)))))..))))).. ( -25.70) >DroSec_CAF1 287980 110 - 1 ACUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU--AACACAAAGUG--------UGUGGAUGCCCGAAUGGAAAAGGGAAAGG ..(((((.......))))).(((((..((((((((((..((((((...........((((.......)))--)((((.....)--------)))))))))))))).)))))..))))).. ( -25.50) >DroSim_CAF1 309324 110 - 1 AAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU--AACACAAAGUG--------UGUGGAUGCCCGAAUGGAAAAGGGAAAGG ..(((((.......))))).(((((..((((((((((..((((((...........((((.......)))--)((((.....)--------)))))))))))))).)))))..))))).. ( -25.70) >DroYak_CAF1 299423 120 - 1 AAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCUAUAGCACAAAGUGUGUAUGUGUGUGGAAGUUCAAAUGGAAAAGGGAAAGG .....(((...(((((...((((((((......))))))))...)))))(((.(((((.(((((((..(....((((.....))))..)..))))))).)..)))).)))..)))..... ( -27.10) >consensus AAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU__AACACAAAGUG________UGUGGAUGCCCGAAUGGAAAAGGGAAAGG .....(((...(((((...((((((((......))))))))...)))))(((.((((.......((((......))))................((....)))))).)))..)))..... (-21.95 = -21.57 + -0.38)



| Location | 5,081,747 – 5,081,865 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5081747 118 - 23771897 AUAUAUUUUCACCCGCAAACUGGUAACAAUUUCCCAUGCAAAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU--AACACAAA ..............(((...(((..........))))))...(((.((...(((((...((((((((......))))))))...)))))...)).)))......((((..--..)))).. ( -18.80) >DroSec_CAF1 288012 118 - 1 AUAUAUUUUCACCCGCAAACUGGUAACAAUUUCCCAUGCAACUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU--AACACAAA ..............(((...((....))......((((.((((((.((...(((((...((((((((......))))))))...)))))...)).)))))).))))))).--........ ( -23.30) >DroSim_CAF1 309356 118 - 1 AUAUAUUUUCACCCGCAAACUGGUAACAAUUUCCCAUGCAAAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU--AACACAAA ..............(((...(((..........))))))...(((.((...(((((...((((((((......))))))))...)))))...)).)))......((((..--..)))).. ( -18.80) >DroYak_CAF1 299463 120 - 1 AUAUAUAUUCACCCGCAAACUGGCAACAAUUUCCAUAGCAAAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCUAUAGCACAAA ..............((....((....))......((((((...((((....(((((...((((((((......))))))))...)))))........)))).....)))))).))..... ( -23.50) >consensus AUAUAUUUUCACCCGCAAACUGGUAACAAUUUCCCAUGCAAAUAACUCAUUUUUAGUUAAUUUCCAAUUUCCUUUGGAAGUGUUCUAAAUUCGAUUUAGUUUUAUGUGCU__AACACAAA ..............(((....((.........))..)))...(((.((...(((((...((((((((......))))))))...)))))...)).)))......((((......)))).. (-17.48 = -17.72 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:06 2006