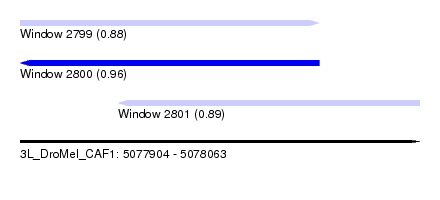
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,077,904 – 5,078,063 |
| Length | 159 |
| Max. P | 0.963369 |

| Location | 5,077,904 – 5,078,023 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5077904 119 + 23771897 ACUUGAUUUUUGAUUAAUCUAAAGCCAGAAAAUA-UUGAUAAAUAUUUGCCAAAUGCAUAUUCGGCUCAGAUGUGGCACAAAAAACCAACUAACAGUAAGAAAAAUUAAUUCGCUGUUUG ......((((((....((((..((((...(((((-((....)))))))((.....))......)))).))))......)))))).......((((((..(((.......))))))))).. ( -18.00) >DroSec_CAF1 284295 119 + 1 AGUUGAUUUUUGAUUAAUCUAAAGCCAGAAAAUA-UUGGUAAAUAUUUGCCAAAUGCAUAUUCGGCCCAGAUGUGGCACAAAAAACCAACUAACAGUGAGAAAAAUUAAUUCGCUGUUUG (((((.((((((....((((...(((.(((.((.-(((((((....)))))))....)).))))))..))))......))))))..)))))(((((((((.........))))))))).. ( -25.80) >DroSim_CAF1 305566 119 + 1 AGUUGAUUUUUGAUUAAUCUAAAGCCAGAAAAUA-UUGGUAAAUAUUUGCCAAAUGCAUAUUCGGCCCAGAUGUGGCACAAAAAACCAACUAACAGUGAGAAAAAUUAAUUCGCUGUUUG (((((.((((((....((((...(((.(((.((.-(((((((....)))))))....)).))))))..))))......))))))..)))))(((((((((.........))))))))).. ( -25.80) >DroEre_CAF1 291364 119 + 1 AGUUGAUUUUUGAUUAAUCUAAAGCCAGAAAGCA-UUGGUAAAUAUUUGCCAAAUGCAUUUUCGACCCAGAUGCGGCACAAAAAACCAACCAACAGUGAGAAAAAUUAAUUCGCUGUUGG .((((.(((((((....))....(((........-(((((((....)))))))..((((((.......)))))))))...))))).))))((((((((((.........)))))))))). ( -30.60) >DroYak_CAF1 295585 120 + 1 AGUUGAUUUUUGGUUAAUCUAAAGCCAGAAAAUAGUUGGUAAAUAUUUGCCAAAUGCAUAUUUGGCCCAGAUGCGGUGCAAAAAACCAACUAAUAGUGAGAAAAAUUAAUUCGCUGUUCG ......(((((((((.......))))))))).((((((((........(((((((....))))))).....(((...)))....))))))))((((((((.........))))))))... ( -30.20) >consensus AGUUGAUUUUUGAUUAAUCUAAAGCCAGAAAAUA_UUGGUAAAUAUUUGCCAAAUGCAUAUUCGGCCCAGAUGUGGCACAAAAAACCAACUAACAGUGAGAAAAAUUAAUUCGCUGUUUG (((((.((((((....((((...(((.(((.....(((((((....))))))).......))))))..))))......))))))..)))))(((((((((.........))))))))).. (-20.76 = -21.44 + 0.68)


| Location | 5,077,904 – 5,078,023 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5077904 119 - 23771897 CAAACAGCGAAUUAAUUUUUCUUACUGUUAGUUGGUUUUUUGUGCCACAUCUGAGCCGAAUAUGCAUUUGGCAAAUAUUUAUCAA-UAUUUUCUGGCUUUAGAUUAAUCAAAAAUCAAGU ((((.(((.(((((((..........))))))).))).)))).((((.......(((((((....)))))))(((((((....))-)))))..))))....((((.......)))).... ( -22.70) >DroSec_CAF1 284295 119 - 1 CAAACAGCGAAUUAAUUUUUCUCACUGUUAGUUGGUUUUUUGUGCCACAUCUGGGCCGAAUAUGCAUUUGGCAAAUAUUUACCAA-UAUUUUCUGGCUUUAGAUUAAUCAAAAAUCAACU ..(((((.((...........)).)))))(((((((((((...((((.......(((((((....)))))))(((((((....))-)))))..))))....((....))))))))))))) ( -26.20) >DroSim_CAF1 305566 119 - 1 CAAACAGCGAAUUAAUUUUUCUCACUGUUAGUUGGUUUUUUGUGCCACAUCUGGGCCGAAUAUGCAUUUGGCAAAUAUUUACCAA-UAUUUUCUGGCUUUAGAUUAAUCAAAAAUCAACU ..(((((.((...........)).)))))(((((((((((...((((.......(((((((....)))))))(((((((....))-)))))..))))....((....))))))))))))) ( -26.20) >DroEre_CAF1 291364 119 - 1 CCAACAGCGAAUUAAUUUUUCUCACUGUUGGUUGGUUUUUUGUGCCGCAUCUGGGUCGAAAAUGCAUUUGGCAAAUAUUUACCAA-UGCUUUCUGGCUUUAGAUUAAUCAAAAAUCAACU .((((((.((...........)).))))))((((((((((.((.....((((((((((.(((.(((((.((..........))))-)))))).)))).))))))..)).)))))))))). ( -29.20) >DroYak_CAF1 295585 120 - 1 CGAACAGCGAAUUAAUUUUUCUCACUAUUAGUUGGUUUUUUGCACCGCAUCUGGGCCAAAUAUGCAUUUGGCAAAUAUUUACCAACUAUUUUCUGGCUUUAGAUUAACCAAAAAUCAACU .....((((((.......))).......((((((((........(((....)))(((((((....)))))))........)))))))).......)))...((((.......)))).... ( -22.20) >consensus CAAACAGCGAAUUAAUUUUUCUCACUGUUAGUUGGUUUUUUGUGCCACAUCUGGGCCGAAUAUGCAUUUGGCAAAUAUUUACCAA_UAUUUUCUGGCUUUAGAUUAAUCAAAAAUCAACU ..(((((.((...........)).)))))(((((((((((.((.....(((((((((((((....))))))).........(((.........)))..))))))..)).))))))))))) (-21.54 = -21.22 + -0.32)


| Location | 5,077,943 – 5,078,063 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -21.82 |
| Energy contribution | -21.94 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5077943 120 - 23771897 CAAACCAAAAUUUGGCAAAUACAAUAAAGCCAAUCGUCGACAAACAGCGAAUUAAUUUUUCUUACUGUUAGUUGGUUUUUUGUGCCACAUCUGAGCCGAAUAUGCAUUUGGCAAAUAUUU ............((((....((((.(((((((((.(....).(((((.(((.......)))...))))).))))))))))))))))).......(((((((....)))))))........ ( -25.00) >DroSec_CAF1 284334 120 - 1 CAAACCAAAAUUUGGCAAAUACAAUAAAGCCAAUCGUCGACAAACAGCGAAUUAAUUUUUCUCACUGUUAGUUGGUUUUUUGUGCCACAUCUGGGCCGAAUAUGCAUUUGGCAAAUAUUU ....(((.....((((....((((.(((((((((.(....).(((((.((...........)).))))).)))))))))))))))))....)))(((((((....)))))))........ ( -28.40) >DroSim_CAF1 305605 120 - 1 CAAACCAAAAUUUGGCAAAUACAAUAAAGCCAAUCGUCGACAAACAGCGAAUUAAUUUUUCUCACUGUUAGUUGGUUUUUUGUGCCACAUCUGGGCCGAAUAUGCAUUUGGCAAAUAUUU ....(((.....((((....((((.(((((((((.(....).(((((.((...........)).))))).)))))))))))))))))....)))(((((((....)))))))........ ( -28.40) >DroEre_CAF1 291403 120 - 1 UAAACCAACAUUGGGCAAAUACUAUAAAGCCAAUCGUCGGCCAACAGCGAAUUAAUUUUUCUCACUGUUGGUUGGUUUUUUGUGCCGCAUCUGGGUCGAAAAUGCAUUUGGCAAAUAUUU ....(((....((((((........(((((((.......((((((((.((...........)).)))))))))))))))...)))).))..)))((((((......))))))........ ( -30.01) >DroYak_CAF1 295625 120 - 1 CAAACCAAAAUUUGGUAAAUACAAUAAAGUCAAUCGACGCCGAACAGCGAAUUAAUUUUUCUCACUAUUAGUUGGUUUUUUGCACCGCAUCUGGGCCAAAUAUGCAUUUGGCAAAUAUUU ....(((.....((((............(((....)))((.(((.(((.(((((((..........))))))).)))))).))))))....)))(((((((....)))))))........ ( -24.40) >consensus CAAACCAAAAUUUGGCAAAUACAAUAAAGCCAAUCGUCGACAAACAGCGAAUUAAUUUUUCUCACUGUUAGUUGGUUUUUUGUGCCACAUCUGGGCCGAAUAUGCAUUUGGCAAAUAUUU ....(((.....(((((....(((.(((((((((.(....).(((((.((...........)).))))).)))))))))))))))))....)))(((((((....)))))))........ (-21.82 = -21.94 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:56 2006