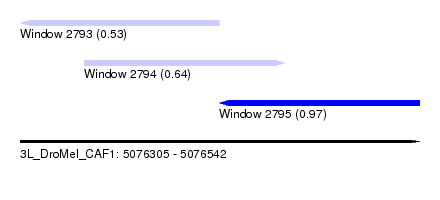
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,076,305 – 5,076,542 |
| Length | 237 |
| Max. P | 0.969257 |

| Location | 5,076,305 – 5,076,423 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.04 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -28.26 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5076305 118 - 23771897 UUCGUGAUGGCAAUAGAGUGCAUCAUUGUGCCACUUGAAAAACUUUGAUUAAAAUCCUUUUGAAGUGCAAACAAAUAGCGCGGGACAAAAGUAUCUGCAGCUUGGAGCUGCCCAGAGA-- ((((.(.(((((...((.....))....)))))).))))...(((((.........((((((..((((.........))))....)))))).....(((((.....))))).))))).-- ( -29.50) >DroSec_CAF1 282701 118 - 1 UUCGAGAUGGCAAUAGAGUGCAUCAUUGUGCCACUUGAAAAACUUUGAUUAAAAUCCUUUUGAAGUGCAAACAAAUAGCGCGGGACAAAAGUAUCUGCAGCUUGGAGCUGCCCAGAGA-- ((((((.(((((...((.....))....)))))))))))...(((((.........((((((..((((.........))))....)))))).....(((((.....))))).))))).-- ( -34.00) >DroSim_CAF1 303979 118 - 1 UUCGAGAUGGCAAUAGAGUGCAUCAUUGUGCCACUUGAAAAACUUUGAUUAAAAUCCUUUUGAAGUGCAAACAAAUAGCGCGGGACAAAAGUAUCUGCAGCUUGGAGCUGCCCAGAGA-- ((((((.(((((...((.....))....)))))))))))...(((((.........((((((..((((.........))))....)))))).....(((((.....))))).))))).-- ( -34.00) >DroEre_CAF1 289754 118 - 1 UUCUAGAUGGCAAUAGAGUGCAUCAUUGUGCCACUUGAAAAACUUUGAUUAAAAUCCUUUUGAAGUGCAAACAAAUAGCGCGGGACAAAAGUAUCUGCAUCUUGGAGCUGCCCAGAGA-- (((.((.(((((...((.....))....))))))).)))...(((((.........((((((..((((.........))))....)))))).....(((.(.....).))).))))).-- ( -23.60) >DroYak_CAF1 293915 120 - 1 UUGGGGAUGGCAAUAGAGUGCAUCAUUGUGCCACUUGAAAAACUUUGAUUAAAAUCCUUUUGAAGUGCAAACAAAUAGCGGGGGACAAAAGUAUCUGCAGCUCGGUGCUGGCCAAAGAGU ....((..((((...((((......((((..(((((.((((...((......))...)))).)))))...))))...(((((..((....)).))))).))))..))))..))....... ( -28.70) >consensus UUCGAGAUGGCAAUAGAGUGCAUCAUUGUGCCACUUGAAAAACUUUGAUUAAAAUCCUUUUGAAGUGCAAACAAAUAGCGCGGGACAAAAGUAUCUGCAGCUUGGAGCUGCCCAGAGA__ ((((((.(((((...((.....))....)))))))))))...(((((.........((((((..((((.........))))....)))))).....(((((.....))))).)))))... (-28.26 = -29.18 + 0.92)

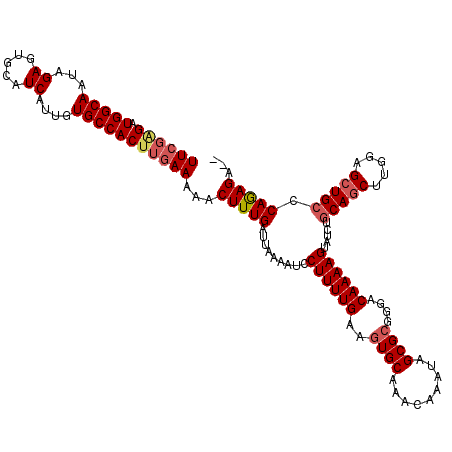
| Location | 5,076,343 – 5,076,462 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.06 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5076343 119 + 23771897 CGCUAUUUGUUUGCACUUCAAAAGGAUUUUAAUCAAAGUUUUUCAAGUGGCACAAUGAUGCACUCUAUUGCCAUCACGAAAGUAGCUGG-UUUCCUUGGCGAAAAAUGCAGACAGAGUGA ((((...((((((((.(((..(((((....(((((....(((((..((((((....((.....))...))))))...)))))....)))-)))))))...)))...)))))))).)))). ( -30.70) >DroSec_CAF1 282739 119 + 1 CGCUAUUUGUUUGCACUUCAAAAGGAUUUUAAUCAAAGUUUUUCAAGUGGCACAAUGAUGCACUCUAUUGCCAUCUCGAAAGUUGCUGG-UUUCCUUGGCGAAAAAUGCAGACAGAGUGA ((((...((((((((.(((..(((((....(((((....(((((.(((((((....((.....))...))))).)).)))))....)))-)))))))...)))...)))))))).)))). ( -31.70) >DroSim_CAF1 304017 119 + 1 CGCUAUUUGUUUGCACUUCAAAAGGAUUUUAAUCAAAGUUUUUCAAGUGGCACAAUGAUGCACUCUAUUGCCAUCUCGAAAGUUGCUGG-UUUCCUUGGCGAAAAAUGCAGACAGAGUGA ((((...((((((((.(((..(((((....(((((....(((((.(((((((....((.....))...))))).)).)))))....)))-)))))))...)))...)))))))).)))). ( -31.70) >DroEre_CAF1 289792 119 + 1 CGCUAUUUGUUUGCACUUCAAAAGGAUUUUAAUCAAAGUUUUUCAAGUGGCACAAUGAUGCACUCUAUUGCCAUCUAGAAAGUUGCUGG-UUUCCUUGGCGAAAACUGAGGACAGAGUGA .((((((((...((.((.....))(((....)))...))....)))))))).........((((((...((((.(.........).)))-).((((..(......)..)))).)))))). ( -27.30) >DroYak_CAF1 293955 120 + 1 CGCUAUUUGUUUGCACUUCAAAAGGAUUUUAAUCAAAGUUUUUCAAGUGGCACAAUGAUGCACUCUAUUGCCAUCCCCAAAGUUGCUGCCUUUCGUUGGCGAAAAACGCAGACAGAGUGA ((((...(((((((..........(((....)))...(((((((.((..((.....((((((......)).))))......))..))(((.......))))))))))))))))).)))). ( -33.10) >consensus CGCUAUUUGUUUGCACUUCAAAAGGAUUUUAAUCAAAGUUUUUCAAGUGGCACAAUGAUGCACUCUAUUGCCAUCUCGAAAGUUGCUGG_UUUCCUUGGCGAAAAAUGCAGACAGAGUGA ((((...(((((((..........(((....)))...(((((((..((((((....((.....))...))))))..........(((((......))))))))))))))))))).)))). (-25.28 = -25.56 + 0.28)



| Location | 5,076,423 – 5,076,542 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.35 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -18.05 |
| Energy contribution | -19.05 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5076423 119 - 23771897 AAAACAAGAGCUGGGUAUAAAAAAAAAAAAAGAAAAACGAAAAUAAUAAUUAAAUCACUGUGAAUAACGGCAGAUAAAUCUCACUCUGUCUGCAUUUUUCGCCAAGGAAACCAGCUACU ........((((((.................(.....)................((...(((((.....(((((((..........)))))))....)))))....))..))))))... ( -21.40) >DroSec_CAF1 282819 116 - 1 AAAACAAGAGCUGGGUAUAAAAA---AUAAAGAAAAAUGAAAAUGAUAAUUAAAUCACUGUGAAUAACGGCAGAUAAAUCUCACUCUGUCUGCAUUUUUCGCCAAGGAAACCAGCAACU .........(((((.........---.................((((......))))..(((((.....(((((((..........)))))))....)))))........))))).... ( -23.60) >DroSim_CAF1 304097 116 - 1 AAAACAAGAGCUGGGUAUAAAAA---AUAAAGAAAAAUGAAAAUGAUAAUUAAAUCACUGUGAAUAACGGCAGAUAAAUCUCACUCUGUCUGCAUUUUUCGCCAAGGAAACCAGCAACU .........(((((.........---.................((((......))))..(((((.....(((((((..........)))))))....)))))........))))).... ( -23.60) >DroEre_CAF1 289872 112 - 1 AAAACAAAAACUGGGUAGAA-------AAAUAAAAAAUGAAAAUGAUAAUUAAAUCACAGUGAAUAACGGCAGAUAAAUCUCACUCUGUCCUCAGUUUUCGCCAAGGAAACCAGCAACU ......(((((((((.....-------................((((......))))...........((((((..........))))))))))))))).((...(....)..)).... ( -18.30) >consensus AAAACAAGAGCUGGGUAUAAAAA___AAAAAGAAAAAUGAAAAUGAUAAUUAAAUCACUGUGAAUAACGGCAGAUAAAUCUCACUCUGUCUGCAUUUUUCGCCAAGGAAACCAGCAACU .........(((((.............................((((......))))..(((((.....(((((((..........)))))))....)))))........))))).... (-18.05 = -19.05 + 1.00)

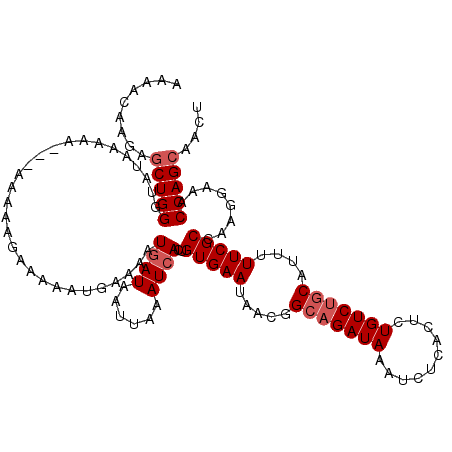
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:50 2006