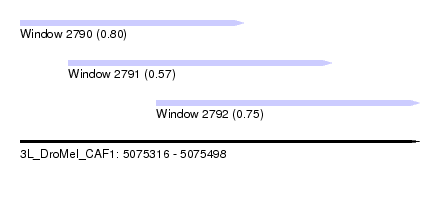
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,075,316 – 5,075,498 |
| Length | 182 |
| Max. P | 0.801430 |

| Location | 5,075,316 – 5,075,418 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5075316 102 + 23771897 AACACCGCAGAGCGGCGG---CGGCGGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUA ..(.((((......))))---.)(((.((....))((((((((...((((((...(((........)))..))))))...)))))))).........)))..... ( -29.00) >DroSec_CAF1 281719 105 + 1 AACACCGCAGAGCGGCGGCAGCGGCGGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUA ....((((...((.((....)).)).)))).....((((((((...((((((...(((........)))..))))))...))))))))................. ( -30.30) >DroSim_CAF1 303007 105 + 1 AACACCGCAGAGCGGCGGCAGCGGCGGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUA ....((((...((.((....)).)).)))).....((((((((...((((((...(((........)))..))))))...))))))))................. ( -30.30) >DroEre_CAF1 288763 96 + 1 AACACCGCAGAGCGGCG---------GCAGCGGCGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGUAUUUA ..(.((((...((....---------)).)))).)((((((((...((((((...(((........)))..))))))...))))))))................. ( -24.00) >consensus AACACCGCAGAGCGGCGG___CGGCGGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUA ......(((..((.((.((....)).)).))....((((((((...((((((...(((........)))..))))))...)))))))).........)))..... (-25.94 = -26.12 + 0.19)



| Location | 5,075,338 – 5,075,458 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5075338 120 + 23771897 GGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCGGGCUCACAGCUCAAAGAACGUGGCGAAC .(((....).((((((((...((((((...(((........)))..))))))...))))))))..........))......(((((.((..((((.....))))...)).).)))).... ( -30.60) >DroSec_CAF1 281744 120 + 1 GGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCUGGCUCACAGCUCAAAGAACGUGGCGAAC .((.((....((((((((...((((((...(((........)))..))))))...))))))))..(((.....((.....((((((....))))))....))...)))...)).)).... ( -32.00) >DroSim_CAF1 303032 120 + 1 GGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCUGGCUCACAGCUCAAAGAACGUGGCGAAC .((.((....((((((((...((((((...(((........)))..))))))...))))))))..(((.....((.....((((((....))))))....))...)))...)).)).... ( -32.00) >DroEre_CAF1 288780 119 + 1 -GCAGCGGCGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGUAUUUAAGCCAGAUCCCGGGCUCACAGCUCAAAGAACGUGGCGAAC -...((.(((((((((((...((((((...(((........)))..))))))...))))))))..((((((.....)))))).........((((.....))))......))).)).... ( -30.80) >DroYak_CAF1 292833 120 + 1 GGCGGCAGCAGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCCGGCUCACAGCUCAAAGAACGUGGCGAAC .((.((....((((((((...((((((...(((........)))..))))))...))))))))..(((.....((.....((((........))))....))...)))...)).)).... ( -28.00) >consensus GGCGGCAACGGCAUCAACAGCAGUAAACAAGCUUCACAUGUGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCGGGCUCACAGCUCAAAGAACGUGGCGAAC .((.((....((((((((...((((((...(((........)))..))))))...))))))))..(((.....((.....((((........))))....))...)))...)).)).... (-27.34 = -27.18 + -0.16)


| Location | 5,075,378 – 5,075,498 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -24.76 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5075378 120 + 23771897 UGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCGGGCUCACAGCUCAAAGAACGUGGCGAACUUUUGCAAUUGUGGGACUAUCAAACAAAACUUUUGCCAAA (((((.......((((((((((((........))))))......((.((((((((.....)))).....(((..((((....))))...))))))))).........))))))))))).. ( -28.91) >DroSec_CAF1 281784 120 + 1 UGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCUGGCUCACAGCUCAAAGAACGUGGCGAACUUUUGCAAUUGUGGGACUAUCAAACAAAACUUUUGCCAAA (((((.........((((((((((........)))))...((((((....))))))..))))).((((......((((....))))..((((..(.....)..))))..))))))))).. ( -28.90) >DroSim_CAF1 303072 120 + 1 UGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCUGGCUCACAGCUCAAAGAACGUGGCGAACUUUUGCAAUUGUGGGACUAUCAAACAAAACUUUUGCCAAA (((((.........((((((((((........)))))...((((((....))))))..))))).((((......((((....))))..((((..(.....)..))))..))))))))).. ( -28.90) >DroEre_CAF1 288819 120 + 1 UGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGUAUUUAAGCCAGAUCCCGGGCUCACAGCUCAAAGAACGUGGCGAACUUUUGCAAUUGUGGGACUAUCAAACAAAACUUUUGCCAAA (((((...........((((((...((((((.....)))))).....((((((((.....)))).....(((..((((....))))...))))))).))))))..........))))).. ( -27.70) >DroYak_CAF1 292873 120 + 1 UGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCCGGCUCACAGCUCAAAGAACGUGGCGAACUUUUGCAAUUGUGGGACUAUCAAACAAAACUUUUGCCAAA (((((.......((((((((((((........))))))...(((((.((...(((.....)))....)).).)))).)))))).....((((..(.....)..))))......))))).. ( -25.00) >consensus UGGCAUUUUAUUAAAGUUGAUGCAACUUAAAAUGCAUUUAAGCCAGAUCCCGGGCUCACAGCUCAAAGAACGUGGCGAACUUUUGCAAUUGUGGGACUAUCAAACAAAACUUUUGCCAAA (((((.......((((((((((((........))))))...(((((.((...(((.....)))....)).).)))).)))))).....((((..(.....)..))))......))))).. (-24.76 = -24.60 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:47 2006