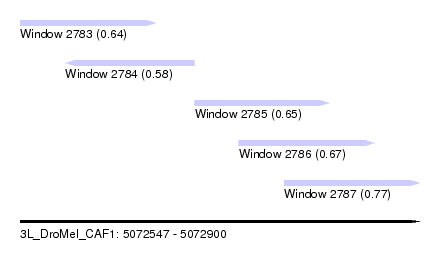
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,072,547 – 5,072,900 |
| Length | 353 |
| Max. P | 0.766405 |

| Location | 5,072,547 – 5,072,667 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5072547 120 + 23771897 AAAACAAAACAAAGCUAAGCGAAGAAUGGAAAAAACAAUCUUUUCAUAAAAUCACGCAAACGCGGCGCUGAAUAAGCUGGCAAACCUAAACAACAAUGCAAUUUUUACUGUUUGUCGCCU ............((((.((((....(((((((........))))))).......(((....))).)))).....))))(((((((.((((.............))))..))))))).... ( -20.42) >DroSec_CAF1 279053 120 + 1 AAAACAAAACAAAGCGAAGCGAAGAAUGGAAAAAACAAUCUUUUCAUAAAAUCACGCAAACGCGGCGCUGAAUAAGCUGGCAAACCUAAACAACAAUGCAAUUUUUACUGUUUGUCGCCU .............(((..(((..(((((((((........)))))))....)).)))...)))((((((.....))).(((((((.((((.............))))..)))))))))). ( -23.12) >DroSim_CAF1 300342 120 + 1 AAAACAAAACAAAGCGAAGCGAAGAAUGGAAAAAACAAUCUUUUCAUAAAAUCACGCAAACGCGGCGCUGAAUAAGCUGGCAAACCUAAACAACAAUGCAAUUUUUACUGUUUGUCGCCU .............(((..(((..(((((((((........)))))))....)).)))...)))((((((.....))).(((((((.((((.............))))..)))))))))). ( -23.12) >DroEre_CAF1 285977 115 + 1 AUAACAAAACAAAGCG-----CAGAAUAGAAAAAACAAUCUUUUCAUAAAAUCACGCAAACGCGGCGCUGAAUAAGCUGGCAAACCUAAACAACAAUGCAAUUUUUACUGUUUGUCGCCU ............((((-----(.(((.(((........))).))).........(((....))))))))......((.(((((((.((((.............))))..))))))))).. ( -21.92) >DroYak_CAF1 289960 110 + 1 AAAGCAAAACAAAGCG----------UAGAAACAACAAUCUUUUCAUAAAAUCACGCAAACGCGGCGCUGAAUAAGCUGGCAAACCUAAACAACAAUGCAAUUUUUACUGUUUGUCGCCU ..(((.......((((----------(.((((.........)))).........(((....))))))))......)))(((((((.((((.............))))..))))))).... ( -18.24) >consensus AAAACAAAACAAAGCGAAGCGAAGAAUGGAAAAAACAAUCUUUUCAUAAAAUCACGCAAACGCGGCGCUGAAUAAGCUGGCAAACCUAAACAACAAUGCAAUUUUUACUGUUUGUCGCCU .......................(((.(((........))).)))..........((....))((((((.....))).(((((((.((((.............))))..)))))))))). (-16.26 = -16.62 + 0.36)



| Location | 5,072,587 – 5,072,701 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5072587 114 - 23771897 CAAAAGAGAGGGGGCGAAGUGUG--UGUG----UGUGAGUAGGCGACAAACAGUAAAAAUUGCAUUGUUGUUUAGGUUUGCCAGCUUAUUCAGCGCCGCGUUUGCGUGAUUUUAUGAAAA .....((((..(.(((((.((((--.(((----(.((((((((((((((.((((....))))..))))).....((....)).)))))))))))))))))))))).)..))))....... ( -32.50) >DroSec_CAF1 279093 116 - 1 CGAAAGAGAGGGGGCGAAGUGUGCGUGUG----UGUGAGUAGGCGACAAACAGUAAAAAUUGCAUUGUUGUUUAGGUUUGCCAGCUUAUUCAGCGCCGCGUUUGCGUGAUUUUAUGAAAA .....((((..(.((((((((.((((.((----.(((((((((((((((.((((....))))..))))))))).((....)).)))))).))))))))).))))).)..))))....... ( -32.60) >DroSim_CAF1 300382 116 - 1 CGAAAGAGAGGGGGCGAAGUGUGCGUGUG----UGUGAGUAGGCGACAAACAGUAAAAAUUGCAUUGUUGUUUAGGUUUGCCAGCUUAUUCAGCGCCGCGUUUGCGUGAUUUUAUGAAAA .....((((..(.((((((((.((((.((----.(((((((((((((((.((((....))))..))))))))).((....)).)))))).))))))))).))))).)..))))....... ( -32.60) >DroEre_CAF1 286012 112 - 1 CGAAAGAGAGGGGGCGAAGUGUGUGUG--------UGAGUAGGCGACAAACAGUAAAAAUUGCAUUGUUGUUUAGGUUUGCCAGCUUAUUCAGCGCCGCGUUUGCGUGAUUUUAUGAAAA .....((((..(.((((((((.((((.--------((((((((((((((.((((....))))..))))).....((....)).)))))))))))))))).))))).)..))))....... ( -31.80) >DroYak_CAF1 289990 120 - 1 GAAAAGAGAGGGGGCGGAGUGUGUGUGUGUGCGUGUGUGUAGGCGACAAACAGUAAAAAUUGCAUUGUUGUUUAGGUUUGCCAGCUUAUUCAGCGCCGCGUUUGCGUGAUUUUAUGAAAA .....((((..(.(((((.((((.((((.((.(((.((.((((((((((.((((....))))..))))))))))((....)).)).))).))))))))))))))).)..))))....... ( -27.90) >consensus CGAAAGAGAGGGGGCGAAGUGUGCGUGUG____UGUGAGUAGGCGACAAACAGUAAAAAUUGCAUUGUUGUUUAGGUUUGCCAGCUUAUUCAGCGCCGCGUUUGCGUGAUUUUAUGAAAA .....((((..(.((((((((.((((.........((((((((((((((.((((....))))..))))).....((....)).)))))))))))))))).))))).)..))))....... (-27.72 = -28.00 + 0.28)


| Location | 5,072,701 – 5,072,820 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.71 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5072701 119 + 23771897 CUAGCAAUCAGCCCGUCUCUUUCGCACCCACCU-ACUACCAUCUCACUCACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAU ..........(((((.(((((((((........-............(((((......))))).....(((((......))))))).)).))))))))..))................... ( -25.00) >DroSec_CAF1 279209 119 + 1 CUCGCAAUCAGCCCGUCUCUUUCGCACCCGCCU-ACUGCCAUCCCACUCACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAU ..........(((((.(((((..((....))..-.(((((......(((((......)))))......((((......)))))))))..))))))))..))................... ( -31.80) >DroSim_CAF1 300498 119 + 1 CUUGCAAUCAGCCUGUCUCUUUCGCACCCACCU-ACCGCCAUCUCACUCACUCCAAUGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAU .((((.....((...........))........-...(((......(((((......)))))((.((((..((((.......))))..)))).))))).))))................. ( -27.90) >DroEre_CAF1 286124 119 + 1 CUCGCAAUCAGCCCAUCUCUGUCACACCCACGC-ACAGCCCUCUCACUCACUCCAACGCGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCGAAACAUUUAAAUCAAAAU .((((.....(((...(((((((........((-(.((.((((.(............).))))..))))).((((.......))))))).)))).))).))))................. ( -24.80) >DroYak_CAF1 290110 120 + 1 CACGCAAUGAGGCCGUCUCUUUCACACCCAUCCAACAGCUAUCUCACACACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAU .......((..((((.(((((((...................(((((..........)))))((....((((......)))).)).)).)))))))))..)).................. ( -27.10) >consensus CUCGCAAUCAGCCCGUCUCUUUCGCACCCACCU_ACAGCCAUCUCACUCACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAU ...((.........((.......))............(((......(((((......)))))((.((((..((((.......))))..)))).))))).))................... (-19.76 = -20.16 + 0.40)

| Location | 5,072,740 – 5,072,860 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5072740 120 + 23771897 AUCUCACUCACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGU ......(((((......)))))(.(((((((((.....(((((((........((....))....(((((......))))).....)))))))((((.....))))))))))))).)... ( -30.00) >DroSec_CAF1 279248 120 + 1 AUCCCACUCACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGU ......(((((......)))))(.(((((((((.....(((((((........((....))....(((((......))))).....)))))))((((.....))))))))))))).)... ( -30.00) >DroSim_CAF1 300537 120 + 1 AUCUCACUCACUCCAAUGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGU ......(((((......)))))(.(((((((((.....(((((((........((....))....(((((......))))).....)))))))((((.....))))))))))))).)... ( -30.60) >DroEre_CAF1 286163 120 + 1 CUCUCACUCACUCCAACGCGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCGAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGU (((.((...((((..((.((((((.((((..((((.......))))..)))).)).(((((....(((((......))))).....)))))...)))).)).))))..)).)))...... ( -31.10) >DroYak_CAF1 290150 120 + 1 AUCUCACACACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGU ........(((......)))(((.(((((((((.....(((((((........((....))....(((((......))))).....)))))))((((.....)))))))))))))))).. ( -29.80) >consensus AUCUCACUCACUCCAACGUGAGGCACUUGCAUGUCACAAUGCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGU ........(((......)))(((.(((((((((.....(((((((........((....))....(((((......))))).....)))))))((((.....)))))))))))))))).. (-28.54 = -28.74 + 0.20)

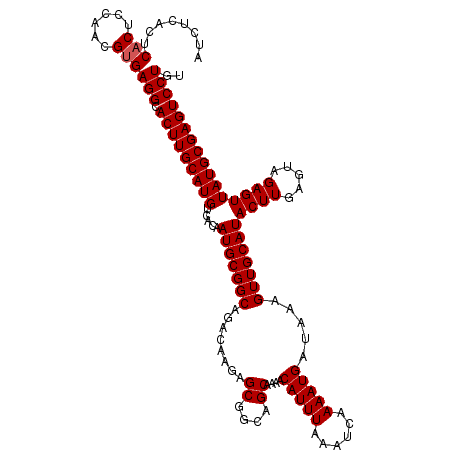

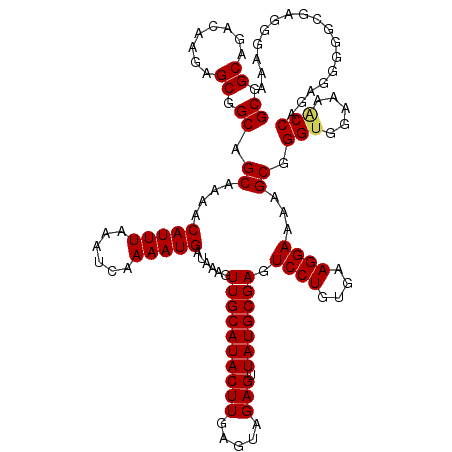
| Location | 5,072,780 – 5,072,900 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5072780 120 + 23771897 GCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGUGAAGGAAAAGCGGGUGGAAAACCAGAGGGGCCGUGGGAGA (((((...((((....(((((....(((((......))))).....)))))...))))..........(((...((((....))))...)))(((.....)))......)))))...... ( -30.90) >DroSec_CAF1 279288 120 + 1 GCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGUGAAGGAAAAGCGGGUGGAAAACCAGAGGGGGCGAGGGAGA (((((........((....))....(((((......))))).....)))))...((((..........(((...((((....))))...)))(((.....)))........))))..... ( -25.20) >DroSim_CAF1 300577 120 + 1 GCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGUGAAGGAAAAGCGGGUGGAAAACCAGAGGGGGCGAGGGAAA (((((........((....))....(((((......))))).....)))))...((((..........(((...((((....))))...)))(((.....)))........))))..... ( -25.20) >DroEre_CAF1 286203 120 + 1 GCGGCAGACAAGAGCGGCAGCGAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGUGAAGGAAAAGCGGGCGGAAAACCGGAGAGGGGGGGGAUAA .(.((...((((....(((((....(((((......))))).....)))))...))))..........(((...((((....))))...))).)).)....((.....)).......... ( -25.40) >DroYak_CAF1 290190 114 + 1 GCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGUGAAGGAAAAGCGGGUGGAAAGCCAGAGGGGUGGA------ ..(((...((((....(((((....(((((......))))).....)))))...)))).......((((.((..((((....))))....)).))))...)))...........------ ( -25.80) >consensus GCGGCAGACAAGAGCGGCAGCAAAACAUUUAAAUCAAAAUGAUAAAGUUGCAUACUUGAGUAGAGUUAUGCGAGUCCUGUGAAGGAAAAGCGGGUGGAAAACCAGAGGGGGCGAGGGAAA ((.((........)).)).((....(((((......)))))......((((((((((.....))).))))))).((((....))))...)).(((.....)))................. (-24.04 = -24.12 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:42 2006