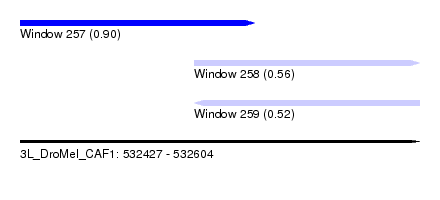
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 532,427 – 532,604 |
| Length | 177 |
| Max. P | 0.901735 |

| Location | 532,427 – 532,531 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -20.15 |
| Consensus MFE | -15.87 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
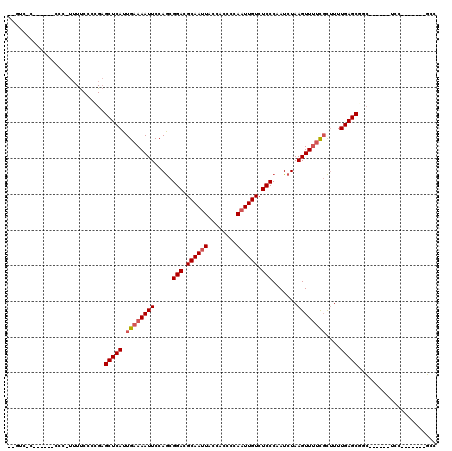
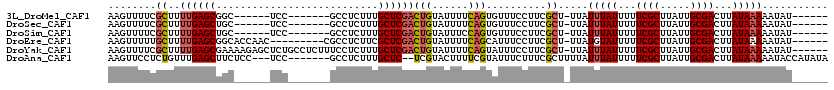
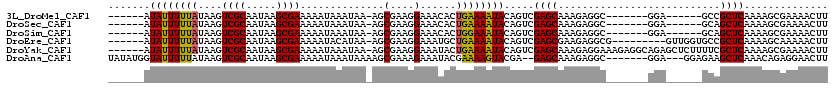
>3L_DroMel_CAF1 532427 104 + 23771897 --GUCCCCCGCACCCC-UGUUCCCCGAGCUCAUUGAAAAUUCCAGCGGACGCAAUUACCACCCCAAUUGUCUCCCAAUCUAAGUUUUCGCUUUUGAGCGGC------UCC-------GCC --.......((....(-(((((...((((.....(((((((..((.(((.((((((........)))))).)))....)).)))))))))))..)))))).------...-------)). ( -19.90) >DroSec_CAF1 6598 98 + 1 --GUCCC------CCC-UUUUCCCCGAGCUCAUUGAAAAUUCCAGCGGACGCAAUUACCACCCCACUUGUCUCCCAAUCUAAGUUUUCGCUUUUGAGCUGC------UCC-------GCC --.....------...-.........((((((.((((((((..((.(((.((((............)))).)))....)).))))))))....))))))..------...-------... ( -15.90) >DroSim_CAF1 7475 98 + 1 --GUCGC------CCC-UUUUCCCCGAGCUCAUUGAAAAUUCCAGCGGACGCAAUUACCACCCCAAUUGUCUCCCAAUCUAAGUUUUCGCUUUUGAGCUGC------UCC-------GCC --.....------...-.........((((((.((((((((..((.(((.((((((........)))))).)))....)).))))))))....))))))..------...-------... ( -19.00) >DroEre_CAF1 6824 96 + 1 --------------CC-UUUUCCCCGAGCUCAUUGAAAAUUCCAGCGGACGCAAUUACCACCCCAAUUGUCUCCCAAUCUAAGUUUUUGCUUUUGAGCGGCACCAAC---------CGCC --------------..-..........(((((............(.(((.((((((........)))))).)))).....((((....)))).)))))(((......---------.))) ( -18.30) >DroYak_CAF1 7027 105 + 1 --------------CC-UUUUCCCCGAGCUCAUUGAAAAUUCCAGCGGACGCAAUUACCACCCCAAUUGUCUCCCAAUCUAAGUUUUCGCUUUUGAGCGAAAAGAGCUCUGCCUCUUUCC --------------..-........((((((.............(.(((.((((((........)))))).))))........((((((((....))))))))))))))........... ( -27.20) >DroAna_CAF1 9947 110 + 1 GCGCCUUUCCCAAGCCAAUUCCCCGGCGCUCAUUGAAAAUUCCAACGGACGCAAUUACCACCCCAAUUGUCUCCCAAUCUAAGUUCCUCUGUUUGAGCUUCUCC---UCC-------GCC (((..........(((........)))((((((((.......))).(((.((((((........)))))).)))...................)))))......---..)-------)). ( -20.60) >consensus __GUC_C______CCC_UUUUCCCCGAGCUCAUUGAAAAUUCCAGCGGACGCAAUUACCACCCCAAUUGUCUCCCAAUCUAAGUUUUCGCUUUUGAGCGGC______UCC_______GCC ...........................(((((.((((((((.....(((.((((((........)))))).))).......))))))))....)))))...................... (-15.87 = -16.32 + 0.44)


| Location | 532,504 – 532,604 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -17.73 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.80 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 532504 100 + 23771897 AAGUUUUCGCUUUUGAGCGGC------UCC-------GCCUCUUUGCUCGACUGUAUUUUCAGUGUUUCCUUCGCU-UUAUUUAUUUUUCGCUUAUUGCGACUUAUAAAAAUAU------ ........((...((((((((------...-------))).....)))))((((......)))).........)).-...(((((...((((.....))))...))))).....------ ( -18.40) >DroSec_CAF1 6669 100 + 1 AAGUUUUCGCUUUUGAGCUGC------UCC-------GCCUCUUUGCUCGACUGUAUUUUCAGUGUUUCCUUCGCU-UUAUUUAUUUUUCGCUUAUUGCGACUUAUAAAAAUAU------ ........((...(((((...------...-------........)))))((((......)))).........)).-...(((((...((((.....))))...))))).....------ ( -16.54) >DroSim_CAF1 7546 100 + 1 AAGUUUUCGCUUUUGAGCUGC------UCC-------GCCUCUUUGCUCGACUGUAUUUCCAGUGUUUCCUUCGCU-UUAUUUAUUUUUCGCUUAUUGCGACUUAUAAAAAUAU------ ........((...(((((...------...-------........)))))((((......)))).........)).-...(((((...((((.....))))...))))).....------ ( -16.04) >DroEre_CAF1 6889 104 + 1 AAGUUUUUGCUUUUGAGCGGCACCAAC---------CGCCUCUUCGCUCGACUGUAUUUUCAGCAUUUCCUUCGCU-UUAUGUAUUUUUCGCUUAUUGCGACUUAUAAAAAUAU------ ..((((((((..(((((((((......---------.))).....))))))(((......)))..........)).-...........((((.....)))).....))))))..------ ( -17.90) >DroYak_CAF1 7092 113 + 1 AAGUUUUCGCUUUUGAGCGAAAAGAGCUCUGCCUCUUUCCUCUUUGCUCGACUGUAUUUUCAGUAUUUCCUUCGCU-UUAUUUAUUUUUCGCUUAUUGCGACUUAUAAAAAUAU------ ......((((...((((((((((((((..................)))).((((......))))............-........))))))))))..)))).............------ ( -25.77) >DroAna_CAF1 10027 108 + 1 AAGUUCCUCUGUUUGAGCUUCUCC---UCC-------GCCUCUUUGCUC--UCGUACUUUUCGUAUUUCUUUCGCUUUUAUUUAUUUUUCGCUUAUUGCGACUUAUAAAAAUACCAUAUA ((((..........((((......---...-------........))))--..((((.....)))).......))))...(((((...((((.....))))...)))))........... ( -11.73) >consensus AAGUUUUCGCUUUUGAGCGGC______UCC_______GCCUCUUUGCUCGACUGUAUUUUCAGUAUUUCCUUCGCU_UUAUUUAUUUUUCGCUUAUUGCGACUUAUAAAAAUAU______ ........((..((((((...........................))))))(((......)))..........)).....(((((...((((.....))))...)))))........... (-11.38 = -11.80 + 0.42)
| Location | 532,504 – 532,604 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -18.98 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.31 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
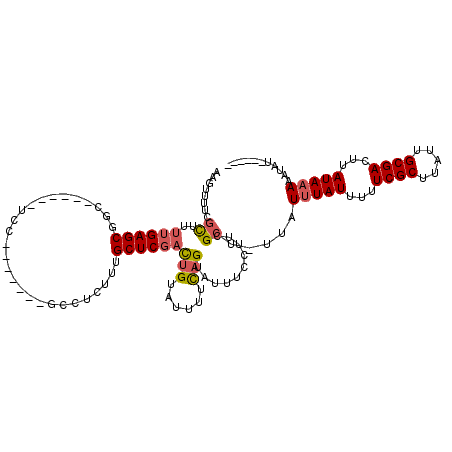
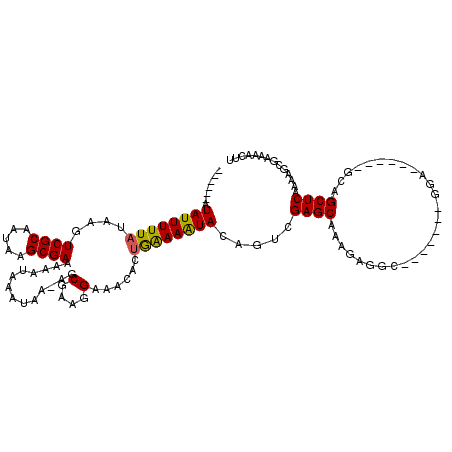
>3L_DroMel_CAF1 532504 100 - 23771897 ------AUAUUUUUAUAAGUCGCAAUAAGCGAAAAAUAAAUAA-AGCGAAGGAAACACUGAAAAUACAGUCGAGCAAAGAGGC-------GGA------GCCGCUCAAAAGCGAAAACUU ------.....(((((...((((.....))))...)))))...-.((...(....)((((......)))).((((.....(((-------...------)))))))....))........ ( -19.10) >DroSec_CAF1 6669 100 - 1 ------AUAUUUUUAUAAGUCGCAAUAAGCGAAAAAUAAAUAA-AGCGAAGGAAACACUGAAAAUACAGUCGAGCAAAGAGGC-------GGA------GCAGCUCAAAAGCGAAAACUU ------.....(((((...((((.....))))...)))))...-.((...(....)((((......)))).((((........-------...------...))))....))........ ( -16.74) >DroSim_CAF1 7546 100 - 1 ------AUAUUUUUAUAAGUCGCAAUAAGCGAAAAAUAAAUAA-AGCGAAGGAAACACUGGAAAUACAGUCGAGCAAAGAGGC-------GGA------GCAGCUCAAAAGCGAAAACUU ------.....(((((...((((.....))))...)))))...-.((...(....)((((......)))).((((........-------...------...))))....))........ ( -18.04) >DroEre_CAF1 6889 104 - 1 ------AUAUUUUUAUAAGUCGCAAUAAGCGAAAAAUACAUAA-AGCGAAGGAAAUGCUGAAAAUACAGUCGAGCGAAGAGGCG---------GUUGGUGCCGCUCAAAAGCAAAAACUU ------.(((((((.....((((.....))))...........-((((.......))))))))))).......((.....((((---------((....)))))).....))........ ( -20.50) >DroYak_CAF1 7092 113 - 1 ------AUAUUUUUAUAAGUCGCAAUAAGCGAAAAAUAAAUAA-AGCGAAGGAAAUACUGAAAAUACAGUCGAGCAAAGAGGAAAGAGGCAGAGCUCUUUUCGCUCAAAAGCGAAAACUU ------.....(((((...((((.....))))...)))))...-.((.........((((......)))).((((.....((((((((......))))))))))))....))........ ( -23.90) >DroAna_CAF1 10027 108 - 1 UAUAUGGUAUUUUUAUAAGUCGCAAUAAGCGAAAAAUAAAUAAAAGCGAAAGAAAUACGAAAAGUACGA--GAGCAAAGAGGC-------GGA---GGAGAAGCUCAAACAGAGGAACUU ......((((((((.....((((.....))))..............(....).......))))))))..--((((........-------...---......)))).............. ( -15.63) >consensus ______AUAUUUUUAUAAGUCGCAAUAAGCGAAAAAUAAAUAA_AGCGAAGGAAACACUGAAAAUACAGUCGAGCAAAGAGGC_______GGA______GCAGCUCAAAAGCGAAAACUU .......((((((((....((((.....))))..............(....)......)))))))).....((((...........................)))).............. (-11.56 = -11.31 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:04 2006