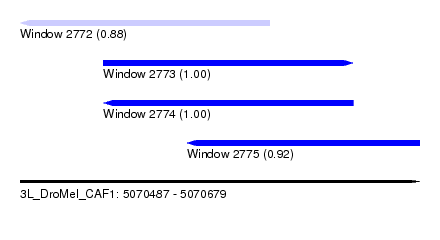
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,070,487 – 5,070,679 |
| Length | 192 |
| Max. P | 0.999891 |

| Location | 5,070,487 – 5,070,607 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -28.76 |
| Energy contribution | -28.36 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5070487 120 - 23771897 ACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGACUUCGCCCUCGGCUUUUUACCCAAAAGUGGAUUCAAUUAA ....((((((((.((((....).))).))))))))((((((.((..((.....(((((((((.......)))))))))......))..)).))))))...(((....))).......... ( -32.20) >DroSec_CAF1 277008 120 - 1 GCUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAAUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGAUUUCGCCCUCGGCUUUUUACCCAAAAGUGGAUUCAAUUAA ((..((((((((.((((....).))).))))))))((((.(((((.((((((..........))))))....)))))...))))))..((.((((((.....)))))).))......... ( -31.50) >DroSim_CAF1 298288 120 - 1 ACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGACUUCGCCCUCGGCUUUUUACCCAAAAGUGGAUUCAAUUAA ....((((((((.((((....).))).))))))))((((((.((..((.....(((((((((.......)))))))))......))..)).))))))...(((....))).......... ( -32.20) >DroEre_CAF1 283914 120 - 1 ACUGAAUGCAUUGCCCUUUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCGUUUUUCCAGGUCUUCGCCCUCGACUUUUUACCCAAAAGUGGAUUCAAUUAA ..(((((.((((.......((((((.((..((...((...((((((((((((..........))))))))...))))..))...))..)).))))))........)))).)))))..... ( -32.56) >DroYak_CAF1 287818 120 - 1 ACUGAAUGCAUUGCCGUGUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGUCUUCGCCCUCGACUUUUUACCCAAAAGUGGAUUCAAUUAA ..((((.(((..(((..(((((((((...(((.......)))...)))))))))((((((((.......)))))))))))..).))..((.((((((.....)))))).))))))..... ( -30.60) >consensus ACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGACUUCGCCCUCGGCUUUUUACCCAAAAGUGGAUUCAAUUAA ..(((((.(...((....((((((((...(.(((.....))).).))))))))(((((((((.......)))))))))......)).....((((((.....))))))).)))))..... (-28.76 = -28.36 + -0.40)



| Location | 5,070,527 – 5,070,647 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5070527 120 + 23771897 UCCUGGAAAAAUGCUCUUUUCUUCCAGUAAGAGCGUUCCACUUUCAAUGCAUUCCACUCUUAUGGCGCAAUGCAUUCAGUUUACUCUUUCGCUCUCUUUUGGCCUUCGAACUGGGCUUUC ..(((((((((......))).))))))..((((((..........(((((((((((......)))...)))))))).............)))))).....(((((.......)))))... ( -27.30) >DroSec_CAF1 277048 120 + 1 UCCUGGAAAAAUGCUCUUUUCUUCCAGUAAGAGCGUUCCAUUUUCAAUGCAUUCCACUCUUAUGGCGCAAUGCAUUCAGCUUACUCUUUCGCUCUCUUUUGGCCUUCGAACAGGGCUUUC ..(((((((((......))).))))))..((((((..........(((((((((((......)))...)))))))).............)))))).....((((((.....))))))... ( -27.50) >DroSim_CAF1 298328 120 + 1 UCCUGGAAAAAUGCUCUUUUCUUCCAGUAAGAGCGUUCCACUUUCAAUGCAUUCCACUCUUAUGGCGCAAUGCAUUCAGUUUACUCUUUCGCUCUCUUUUGGCCUUCGAACAGGGCUUUC ..(((((((((......))).))))))..((((((..........(((((((((((......)))...)))))))).............)))))).....((((((.....))))))... ( -27.50) >DroEre_CAF1 283954 120 + 1 ACCUGGAAAAACGCUCUUUUCUUCCAGUAAGAGCGUUCCACUUUCAAUGCAUUCCACUCUUAAAGGGCAAUGCAUUCAGUUUACUCUUUCGCUCUCUUUUGGCCUUCAAACUGGGGAUUC .((((((...((((((((..........)))))))))))......((((((((((.((.....)))).))))))))((((((........(((.......)))....))))))))).... ( -29.00) >DroYak_CAF1 287858 120 + 1 ACCUGGAAAAAUGCUCUUUUCUUCCAGUAAGAGCGUUCCACUUUCAAUGCAUUCCACUCUUACACGGCAAUGCAUUCAGUUUACUCUUUCGCUCUCUUUUGGCCUUCAAACUGGGCUUUC ..(((((((((......))).))))))..((((((..........((((((((((..........)).)))))))).............)))))).....(((((.......)))))... ( -27.80) >consensus UCCUGGAAAAAUGCUCUUUUCUUCCAGUAAGAGCGUUCCACUUUCAAUGCAUUCCACUCUUAUGGCGCAAUGCAUUCAGUUUACUCUUUCGCUCUCUUUUGGCCUUCGAACUGGGCUUUC ..(((((((((......))).))))))..((((((..........((((((((.(.((.....)).).)))))))).............)))))).....(((((.......)))))... (-24.10 = -24.46 + 0.36)



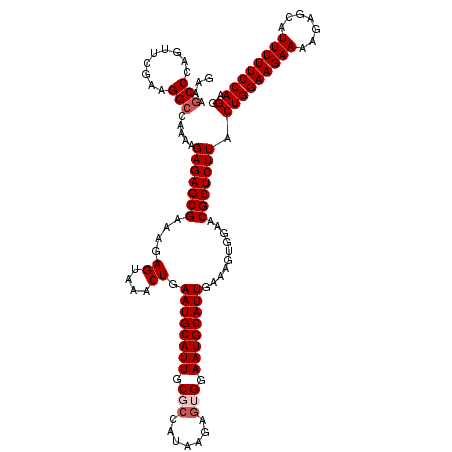
| Location | 5,070,527 – 5,070,647 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -31.16 |
| Energy contribution | -31.96 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5070527 120 - 23771897 GAAAGCCCAGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGA ....(((.........))).....(((((((....((....)).((((((((.((((....).))).))))))))..........))))))).(((((((((.......))))))))).. ( -34.80) >DroSec_CAF1 277048 120 - 1 GAAAGCCCUGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAGCUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAAUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGA ....(.(((......))).)....(((((((....((....)).((((((((.((((....).))).))))))))..........))))))).(((((((((.......))))))))).. ( -35.70) >DroSim_CAF1 298328 120 - 1 GAAAGCCCUGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGA ....(.(((......))).)....(((((((....((....)).((((((((.((((....).))).))))))))..........))))))).(((((((((.......))))))))).. ( -35.40) >DroEre_CAF1 283954 120 - 1 GAAUCCCCAGUUUGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCCCUUUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCGUUUUUCCAGGU ..........((((.....)))).(((((((....((....)).((((((((.(((((...))).))))))))))..........))))))).(((((((((.......))))))))).. ( -31.10) >DroYak_CAF1 287858 120 - 1 GAAAGCCCAGUUUGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCCGUGUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGU ....(((.........))).....(((((((....((....)).((((((((.(((........)))))))))))..........))))))).(((((((((.......))))))))).. ( -32.60) >consensus GAAAGCCCAGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAGUGGAACGCUCUUACUGGAAGAAAAGAGCAUUUUUCCAGGA ....(((.........))).....(((((((....((....)).((((((((.(((.......))).))))))))..........))))))).(((((((((.......))))))))).. (-31.16 = -31.96 + 0.80)


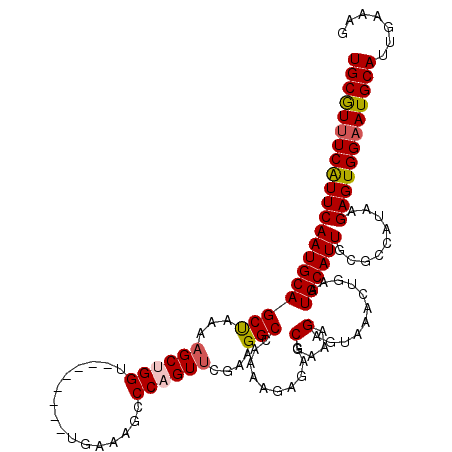
| Location | 5,070,567 – 5,070,679 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5070567 112 - 23771897 UGCGUUUCAUUCAAUGCAGCUAAAAGCUGGU--------UGAAAGCCCAGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAG ((((((((((((............((((((.--------(....).))))))....((((((.......(....).(((.......))).))).)))....))))))))))))....... ( -30.30) >DroSec_CAF1 277088 112 - 1 UGCGUCUCAUUCAAUGCAGCUAAAAGCUGGU--------UGAAAGCCCUGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAGCUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAA .((.((((.((((((.((((.....))))))--------)))).(.(((......))).)....))))))............(.((((((((.((((....).))).)))))))).)... ( -32.90) >DroSim_CAF1 298368 112 - 1 UGCGUUUCAUUCAAUGCAGCUAAAAGCUGGU--------UGAAAGCCCUGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAG .((.((((.((((((.((((.....))))))--------)))).(.(((......))).)....))))))............(.((((((((.((((....).))).)))))))).)... ( -30.30) >DroEre_CAF1 283994 112 - 1 UGCGUUUCAUUCAAUGCAGCCAAAAGCUGGU--------UGAAUCCCCAGUUUGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCCCUUUAAGAGUGGAAUGCAUUGAAAG ((((((((((((......(((..(((((((.--------.......)))))))...)))....((((.((((....(((.......))).)))).))))..))))))))))))....... ( -32.80) >DroYak_CAF1 287898 120 - 1 UGCAUUUCGUUCAAUGCAGCCAAAUGCUGGUUGACUGGUUGAAAGCCCAGUUUGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCCGUGUAAGAGUGGAAUGCAUUGAAAG (((.((((((((...(((......)))(((((((((((.(....).))))))....))))).....))))))))..)))...(.((((((((.(((........))))))))))).)... ( -35.60) >consensus UGCGUUUCAUUCAAUGCAGCUAAAAGCUGGU________UGAAAGCCCAGUUCGAAGGCCAAAAGAGAGCGAAAGAGUAAACUGAAUGCAUUGCGCCAUAAGAGUGGAAUGCAUUGAAAG (((((((((((((((((((((...((((((................))))))....)))..........(....)...........)))))).........))))))))))))....... (-24.43 = -24.67 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:29 2006