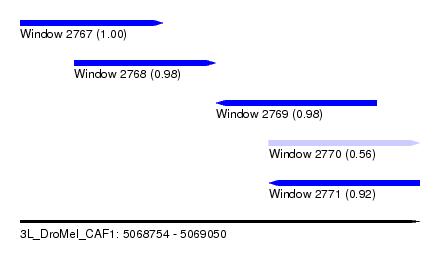
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,068,754 – 5,069,050 |
| Length | 296 |
| Max. P | 0.999561 |

| Location | 5,068,754 – 5,068,860 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -34.36 |
| Energy contribution | -34.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5068754 106 + 23771897 AUCGGAAAAGCAGGCGGCCAGCCCCAAAACUGCUGUCUGCUGGUA--------------ACUGGUAACUGGUAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGAAGCACAGC (((((...((((((((((.((........))))))))))))....--------------.)))))..(((((....)))))..((((((((...........)))))))).......... ( -35.70) >DroSec_CAF1 275307 106 + 1 AUCGGAAAAGCAGGCGGCCAGCCCCAAAACUGCUGGCUGCUGGUG--------------ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGC .........((.((((((((((.........)))))))))).(((--------------.((((((...(....)))))))(.((((((((...........)))))))).)..))).)) ( -40.50) >DroSim_CAF1 296584 106 + 1 AUCGGAAAAGCAGGCGGCCAGCCCCAAAACUGCUGGCUGCUGGUG--------------ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGC .........((.((((((((((.........)))))))))).(((--------------.((((((...(....)))))))(.((((((((...........)))))))).)..))).)) ( -40.50) >DroEre_CAF1 282184 120 + 1 AUCGGAAAAGCAGGCGGCCAGCCCCAAAACUGCUGGCUGCUGGCUGCUGGCUGCUGGUAACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGC ........(((.((((((((((.........)))))))))).)))((((((((((((((...(....).(....)))))))).((((((((...........))))))))..))).)))) ( -51.80) >DroYak_CAF1 286047 106 + 1 AUCGGAAAAGCAGGCGUCCAGCCCCAAAACUGCUGGCUACUGGCU--------------ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCAUAGC ...(((...(....).))).((.((.....((((((((((..(..--------------.)..)))...(....).)))))))((((((((...........)))))))))).))..... ( -34.80) >consensus AUCGGAAAAGCAGGCGGCCAGCCCCAAAACUGCUGGCUGCUGGUG______________ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGC ..((.....((.((((((((((.........)))))))))).))................((((((...(....)))))))))((((((((...........)))))))).......... (-34.36 = -34.72 + 0.36)

| Location | 5,068,794 – 5,068,899 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.17 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -29.24 |
| Energy contribution | -29.16 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5068794 105 + 23771897 UGGUA--------------ACUGGUAACUGGUAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGAAGCACAGCCAACUAACUCAGCCAUUAUCCCCAUGGGUAGUUGC-CGGG .....--------------........((((((((((((.((.((((((((...........))))))))...))..................(((.......))))))))))))-))). ( -31.90) >DroSec_CAF1 275347 105 + 1 UGGUG--------------ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGCCAACUAACUCAGCCAUUAUCCCCAUGGGUAGUUGC-CGGG ((((.--------------.((((((...(....)))))))(.((((((((...........)))))))).)......)))).....(((.((.(((((((....))))))).))-.))) ( -33.50) >DroSim_CAF1 296624 105 + 1 UGGUG--------------ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGCCAACUAACUCAGCCAUUAUCCCCAUGGGUAGUUGC-CGGG ((((.--------------.((((((...(....)))))))(.((((((((...........)))))))).)......)))).....(((.((.(((((((....))))))).))-.))) ( -33.50) >DroEre_CAF1 282224 119 + 1 UGGCUGCUGGCUGCUGGUAACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGCCAACUAACUCAGCCAUUAUCCCCAUGGGUAGCUGC-CAGG ((((.(((((((((((((..((((((...(....)))))))(.((((((((...........)))))))).).)).))))..........))))).(((((....))))))).))-)).. ( -42.00) >DroYak_CAF1 286087 106 + 1 UGGCU--------------ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCAUAGCCAACUAACUCAGCCAUUAUCCCCAUGGGUAGCUGCCCAGG (((((--------------(((((((...(....)))))))(.((((((((...........)))))))).)....))))))...................((.(((((....))))))) ( -34.90) >consensus UGGUG______________ACUGGUAACUGGAAACUGCCAGCGGAACAGAUGCAAUCAUAAAAUCUGUUCGGAGCACAGCCAACUAACUCAGCCAUUAUCCCCAUGGGUAGUUGC_CGGG ((((................((((((...(....)))))))..((((((((...........))))))))........)))).....((..((.(((((((....))))))).))..)). (-29.24 = -29.16 + -0.08)


| Location | 5,068,899 – 5,069,018 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -31.88 |
| Energy contribution | -32.56 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5068899 119 - 23771897 AAACGGAAGAACUUGUUCCCUGCCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAUUUCUCGGUCACCUCGAACCCGACUGCC-CCCAUCCUUCCG ...((((((.......(((((((((((((((...((....)))))))))(((((((....)).)))))..).))))))).....(((((...........)))))..-......)))))) ( -43.30) >DroSec_CAF1 275452 117 - 1 A-ACCGAAGACCUUGUUCACUGGCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAUUUCUCGGCCACCUCGAACCCGACUGCC--CCAUCCUUCCG .-...((((.((((((((((((.((((((((...((....)))))))))).))))((((.....))))..)))))))).......(((....(((....)))..)))--.....)))).. ( -39.40) >DroSim_CAF1 296729 118 - 1 A-ACCGAAGACCUUGUUCACUGGCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAUUUCUCGGCCACCUCGAACCCGACUGCC-CCCAUCCUUCCG .-...((((.((((((((((((.((((((((...((....)))))))))).))))((((.....))))..)))))))).......(((....(((....)))..)))-......)))).. ( -39.40) >DroEre_CAF1 282343 116 - 1 A-ACCGAAGACCUUGUCCACUGCCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAUUUCUCGGCCACCUCAAACCCGACUGC---CCAUCCUUCCG .-...((((...(((.((((((.((((((((...((....)))))))))).)))))))))...............(((.....((((...........))))...)---))...)))).. ( -35.20) >DroYak_CAF1 286193 119 - 1 A-ACCGAAGACCUUGUUCGCUGCCGACUCAAACUAGUUGUCCUUGAGUAGACAGUGGCAACAUUUGUCAUGAGCAGGGAUUUCUCGGCCAGCUCAAGCCCGAGUGCCACCCAUCCCUCCC .-...((((.(((((((((......((((((((.....))..)))))).(((((((....)).))))).))))))))).))))..(((..((((......)))))))............. ( -36.80) >consensus A_ACCGAAGACCUUGUUCACUGCCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAUUUCUCGGCCACCUCGAACCCGACUGCC_CCCAUCCUUCCG .....((((.((((((((((((.((((((((((.....))..)))))))).))))((((.....))))..)))))))).))))((((...........)))).................. (-31.88 = -32.56 + 0.68)

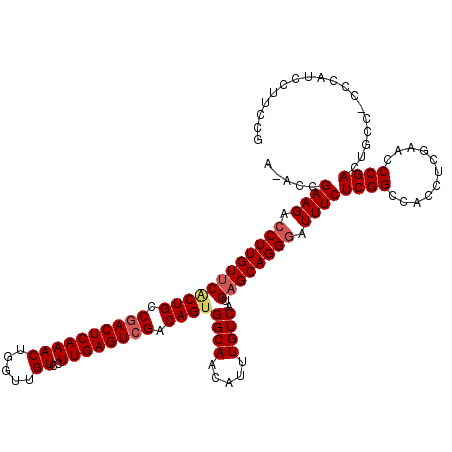
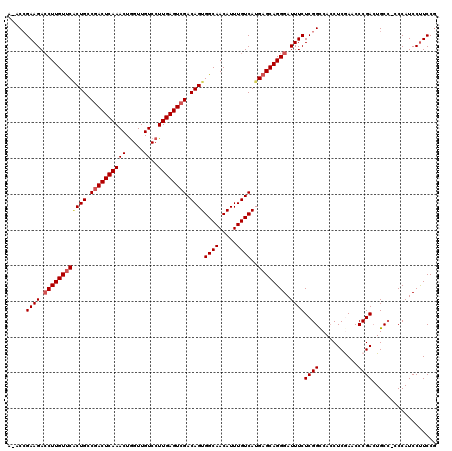
| Location | 5,068,938 – 5,069,050 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.38 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -23.08 |
| Energy contribution | -24.36 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5068938 112 + 23771897 AUCCCUGCUCAUGACAAAUGUUGCCACUGUCGACUCAAGGACAACCAGUUUGAGUCGGCAGGGAACAAGUUCUUCCGUUUUCCGUUCC-AAAGAAGUGACC-------CAGUAAAUCUGA ...................((..(..((((((((((((((....))...))))))))))))(((((.................)))))-......)..)).-------(((.....))). ( -29.33) >DroSec_CAF1 275490 112 + 1 AUCCCUGCUCAUGACAAAUGUUGCCACUGUCGACUCAAGGACAACCAGUUUGAGUCGCCAGUGAACAAGGUCUUCGGU-UUCCGUUCCGAAAGAAGUGACC-------CAGUAAAUCUGA ....(((.(((((((...((((..(((((.((((((((((....))...)))))))).)))))))))..)))(((((.-.......)))))....))))..-------)))......... ( -32.40) >DroSim_CAF1 296768 112 + 1 AUCCCUGCUCAUGACAAAUGUUGCCACUGUCGACUCAAGGACAACCAGUUUGAGUCGCCAGUGAACAAGGUCUUCGGU-UUCCGUUCGAAAAGAAGUGACC-------CAGUAAAUCUGA ....(((.(((((((...((((..(((((.((((((((((....))...)))))))).)))))))))..)))(((((.-......))))).....))))..-------)))......... ( -29.90) >DroEre_CAF1 282380 110 + 1 AUCCCUGCUCAUGACAAAUGUUGCCACUGUCGACUCAAGGACAACCAGUUUGAGUCGGCAGUGGACAAGGUCUUCGGU-UUCCGUUCUAAAC---------GCAGGGACAUUAAAUCUGA .(((((((............((((((((((((((((((((....))...))))))))))))))).)))((.(....).-..)).........---------)))))))............ ( -45.40) >DroYak_CAF1 286233 119 + 1 AUCCCUGCUCAUGACAAAUGUUGCCACUGUCUACUCAAGGACAACUAGUUUGAGUCGGCAGCGAACAAGGUCUUCGGU-UUCCGUUUCAAACAAAGUGACCGCAAUGACAGUAAAUCUGA ....(((.((((..(...(((((((..(((((......)))))(((......))).))))))).....((((....((-((.......)))).....)))))..)))))))......... ( -27.60) >consensus AUCCCUGCUCAUGACAAAUGUUGCCACUGUCGACUCAAGGACAACCAGUUUGAGUCGGCAGUGAACAAGGUCUUCGGU_UUCCGUUCCAAAAGAAGUGACC_______CAGUAAAUCUGA ............(((...((((..((((((((((((((((....))...))))))))))))))))))..)))..(((....)))........................(((.....))). (-23.08 = -24.36 + 1.28)

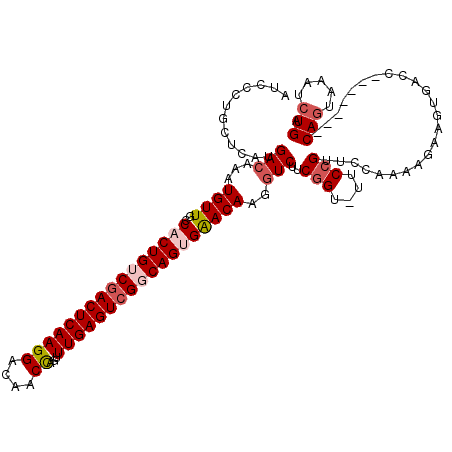
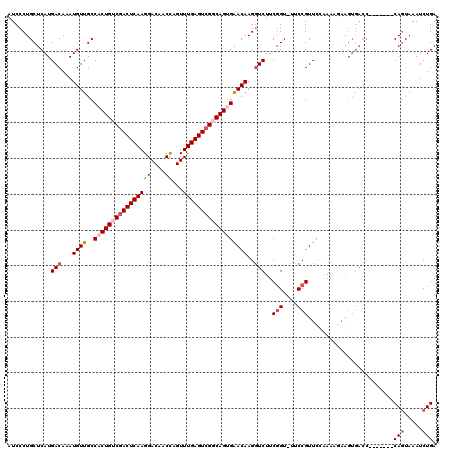
| Location | 5,068,938 – 5,069,050 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.38 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5068938 112 - 23771897 UCAGAUUUACUG-------GGUCACUUCUUU-GGAACGGAAAACGGAAGAACUUGUUCCCUGCCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAU .........(((-------..(((......)-))..)))....(....).......(((((((((((((((...((....)))))))))(((((((....)).)))))..).))))))). ( -37.20) >DroSec_CAF1 275490 112 - 1 UCAGAUUUACUG-------GGUCACUUCUUUCGGAACGGAA-ACCGAAGACCUUGUUCACUGGCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAU ((((.....)))-------)........((((((.......-.)))))).((((((((((((.((((((((...((....)))))))))).))))((((.....))))..)))))))).. ( -38.30) >DroSim_CAF1 296768 112 - 1 UCAGAUUUACUG-------GGUCACUUCUUUUCGAACGGAA-ACCGAAGACCUUGUUCACUGGCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAU ((((.....)))-------)....(((((((((....))))-)..)))).((((((((((((.((((((((...((....)))))))))).))))((((.....))))..)))))))).. ( -37.50) >DroEre_CAF1 282380 110 - 1 UCAGAUUUAAUGUCCCUGC---------GUUUAGAACGGAA-ACCGAAGACCUUGUCCACUGCCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAU ...........((((((((---------........(((..-.)))..(((..(((((((((.((((((((...((....)))))))))).))))))..)))...)))....)))))))) ( -43.20) >DroYak_CAF1 286233 119 - 1 UCAGAUUUACUGUCAUUGCGGUCACUUUGUUUGAAACGGAA-ACCGAAGACCUUGUUCGCUGCCGACUCAAACUAGUUGUCCUUGAGUAGACAGUGGCAACAUUUGUCAUGAGCAGGGAU ((((((..(((((....)))))......))))))..(((..-.)))....(((((((((......((((((((.....))..)))))).(((((((....)).))))).))))))))).. ( -32.90) >consensus UCAGAUUUACUG_______GGUCACUUCUUUUGGAACGGAA_ACCGAAGACCUUGUUCACUGCCGACUCAAACUGGUUGUCCUUGAGUCGACAGUGGCAACAUUUGUCAUGAGCAGGGAU ....................................(((....)))....((((((((((((.((((((((((.....))..)))))))).))))((((.....))))..)))))))).. (-27.98 = -28.86 + 0.88)

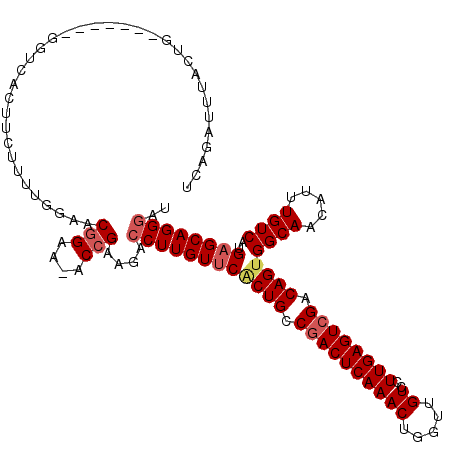
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:25 2006