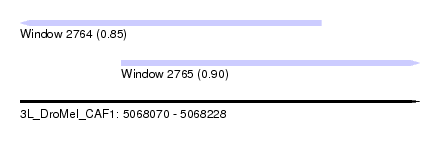
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,068,070 – 5,068,228 |
| Length | 158 |
| Max. P | 0.897272 |

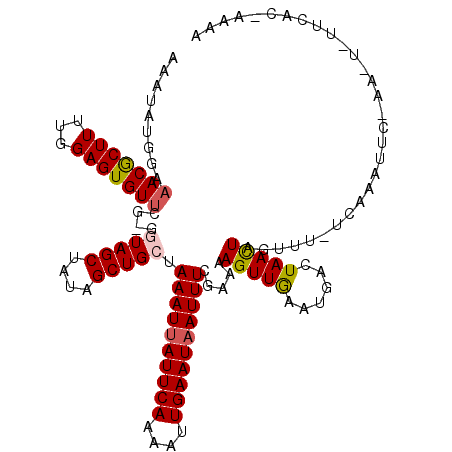
| Location | 5,068,070 – 5,068,189 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.38 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -18.14 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5068070 119 - 23771897 UCAACUUUUCGAAAUAAUUCAAUUUUGAAUAAUUUAGCAGCUAUAGCUACC-CGAACACUCCAAAAGCGUUUUCAUAUUUACAUAUGUAAACAAAAUCAUGAGGGGAAAUUACAGGAGAA .......((((((((.(((((....))))).))))...(((....)))...-))))..((((......((((.(((((....))))).))))....((.......)).......)))).. ( -18.00) >DroSec_CAF1 274592 120 - 1 UCAACUUUUCGAAAUUAUUCAAUUUUGAAUAAUUUAGCAGCUAUAGCUAGCCCGAACACUCCAAAAGCGUUUCCAUAUUUACAUAUGUAAACAAAAUCAUGAGGGGAAAUUAGAGGAGAA .......((((((((((((((....)))))))))).(((((....))).)).))))..((((......((((((...(((((....)))))((......))...))))))....)))).. ( -25.50) >DroSim_CAF1 295923 119 - 1 UCAACUUUUCGAAAUUAUUCAAUUUUGAAUAAUUUAGCAGCUAUAGCUAGC-CGAACACUCCAAAAGCGUUUCCAUAUUUACAUAUGUAAACAAAAUCAUGAGGGGAAAUUAGAGGAGAA .......((((((((((((((....)))))))))).(((((....))).))-))))..((((......((((((...(((((....)))))((......))...))))))....)))).. ( -25.50) >DroEre_CAF1 281510 119 - 1 CUAACUUUUCGAAAUUAUUCAAUUUUGAAUAAUUUAGCAGCUACAGCUACA-ACAACACUCUAAAAGUGUUUCCAUAUUUACAUAUGUAAACAAAAUCAUGAGGGGACAUUACAGGAGAA ...........((((((((((....)))))))))).(.(((....))).).-......((((...(((((..((...(((((....)))))((......)).))..)))))...)))).. ( -24.90) >DroYak_CAF1 285368 116 - 1 CUAACUUUUCGAAAUUAUUCAAUUUUGAAUAAUUUAGCAGCUUCAGCUACG----ACACUCUAAAAGCGUUUCCAUAUUUACAUAUGUAAACAAAAUCAUGAGGGGACAUUACAGGAGUA ........(((((((((((((....))))))))))...(((....))).))----).(((((......((..((...(((((....)))))((......)).))..))......))))). ( -23.60) >consensus UCAACUUUUCGAAAUUAUUCAAUUUUGAAUAAUUUAGCAGCUAUAGCUACC_CGAACACUCCAAAAGCGUUUCCAUAUUUACAUAUGUAAACAAAAUCAUGAGGGGAAAUUACAGGAGAA ...........((((((((((....))))))))))...(((....)))..........((((.......(((((...(((((....)))))((......)).))))).......)))).. (-18.14 = -17.94 + -0.20)

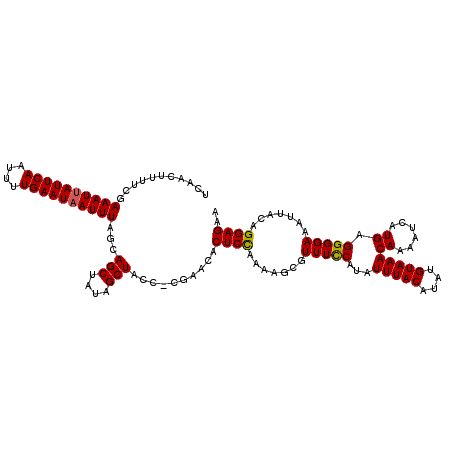
| Location | 5,068,110 – 5,068,228 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5068110 118 + 23771897 AAAUAUGAAAACGCUUUUGGAGUGUUCG-GGUAGCUAUAGCUGCUAAAUUAUUCAAAAUUGAAUUAUUUCGAAAAGUUGAAUGACUAAUUAGUUU-UCAGAUUCAAAUUAUUCACUAAAA .....(((((((....((((........-((((((....))))))...((((((((..(((((....)))))....))))))))))))...))))-)))..................... ( -25.40) >DroSec_CAF1 274632 107 + 1 AAAUAUGGAAACGCUUUUGGAGUGUUCGGGCUAGCUAUAGCUGCUAAAUUAUUCAAAAUUGAAUAAUUUCGAAAAGUUGAAUGACUAACUAGUUUUUUAAAUUG-------------AAA ......(....)((((((.((.......(((.(((....)))))).(((((((((....))))))))))).))))))((((.((((....)))).)))).....-------------... ( -21.80) >DroSim_CAF1 295963 105 + 1 AAAUAUGGAAACGCUUUUGGAGUGUUCG-GCUAGCUAUAGCUGCUAAAUUAUUCAAAAUUGAAUAAUUUCGAAAAGUUGAAUGACUAACUAGUUU-UCAAAUUC-------------AAA .....(((((((....((((..((((((-((((((....))).(.((((((((((....)))))))))).)...))))))))).))))...))))-))).....-------------... ( -24.10) >DroEre_CAF1 281550 118 + 1 AAAUAUGGAAACACUUUUAGAGUGUUGU-UGUAGCUGUAGCUGCUAAAUUAUUCAAAAUUGAAUAAUUUCGAAAAGUUAGAUGACUAACUAGCUU-UUGAAUUUUAAAUUUUCACCAAAC .....(((.(((((((...)))))))..-.(((((....))))).((((((((((....))))))))))..(((((((((........)))))))-))................)))... ( -27.10) >DroYak_CAF1 285408 114 + 1 AAAUAUGGAAACGCUUUUAGAGUGU----CGUAGCUGAAGCUGCUAAAUUAUUCAAAAUUGAAUAAUUUCGAAAAGUUAGUUGACUAACUAGGUU-GCGAAUUCGAAUUUUUCACUAAA- .....(((((((((((...)))))(----(((((((.......(.((((((((((....)))))))))).)...((((((....)))))).))))-))))........)))))).....- ( -26.60) >consensus AAAUAUGGAAACGCUUUUGGAGUGUUCG_GGUAGCUAUAGCUGCUAAAUUAUUCAAAAUUGAAUAAUUUCGAAAAGUUGAAUGACUAACUAGUUU_UCAAAUUC_AA_U_UUCAC_AAAA .........(((((((...)))))))....(((((....))))).((((((((((....)))))))))).....(((((......))))).............................. (-15.88 = -16.12 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:19 2006