| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 532,045 – 532,141 |
| Length | 96 |
| Max. P | 0.789293 |

| Location | 532,045 – 532,141 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.35 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -10.02 |
| Energy contribution | -10.05 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
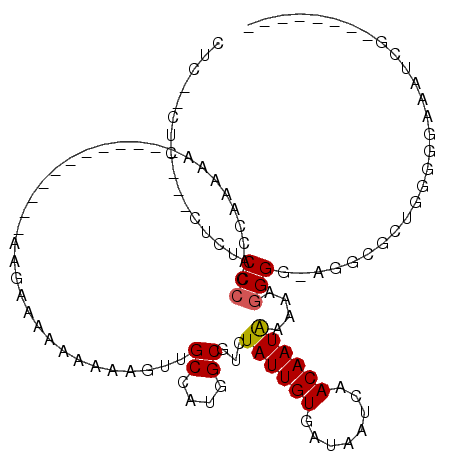
>3L_DroMel_CAF1 532045 96 - 23771897 CU---CUCA--CCAACCAAACCAAAAA----------UAAAGAAGAAAAAAGUUGCCAUGGCGUCUAUUGUGAUAAUCAACAAUAAAAAGGGG-AGGCGCUGGGGGAAAUCG-------- .(---(((.--(((.((...((.....----------.................((....))...((((((........))))))....))..-.))...))))))).....-------- ( -17.10) >DroSec_CAF1 6222 91 - 1 CUCCUCUCU--CUUUCCCGACCGACA-----------------AAAAAAAAGUUGCCAUGGCGUCUAUUGUGAUAAUCAACAAUAAAAAGGGA-AGGCGC-GGAGGAAAUCG-------- .(((((((.--((((((((((.(((.-----------------........)))((....)))))((((((........))))))....))))-))).).-)))))).....-------- ( -28.20) >DroSim_CAF1 7096 92 - 1 CUCCUCUCUCUCUCUCCCUACCCACA-----------------AA--AAAAGUUGCCAUGGCGUCUAUUGUGAUAAUCAACAAUAAAAAGGGG-AGGCGCUGGGGGAAAUCG-------- .((((((..(.(.((((((.......-----------------..--.......((....))...((((((........))))))....))))-))).)..)))))).....-------- ( -25.00) >DroEre_CAF1 6446 89 - 1 CUGU---------CUCCCCACCAAAAA------------AAGAAGAAAAAAGUUGCCAUGGCGUCUAUUGUGAUAAUCAACAAUAAAAAGGGG--GGCGCGGGGGGGAAUCG-------- ((((---------((((((........------------...............((....))...((((((........))))))....))))--)).))))..........-------- ( -22.00) >DroYak_CAF1 6636 111 - 1 CUCUCCUCCCU-UCACCCCACCAAAAAAAGAGGGGAAAAAAGAAGAAAAAAGUUGCCAUGGCGUCUAUUGUGAUAAUCAACAAUAAAAAGGGGGAGGCGCCGGGGGAAAACG-------- ...((((((((-((.((((.(........).))))......))))..............((((((((((((........)))))..........))))))))))))).....-------- ( -29.60) >DroAna_CAF1 9503 98 - 1 CU---------CUU-CCCCGUUGGAAA----------U-GGAAAAAAAAAAGUUGCCAUGGCGUCUAUUGUGAUUAUCAACAAUGAAAAGGGG-AAGGGGAAAGAGAAAAGGGGGGGCGG ((---------(((-((((....(..(----------(-((...((......)).))))..)...((((((........))))))....))))-)))))..................... ( -24.10) >consensus CUC__CUC___CUCUCCCCACCAAAAA____________AAGAAAAAAAAAGUUGCCAUGGCGUCUAUUGUGAUAAUCAACAAUAAAAAGGGG_AGGCGCUGGGGGAAAUCG________ ...............(((....................................((....))...((((((........))))))....)))............................ (-10.02 = -10.05 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:54 2006