
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
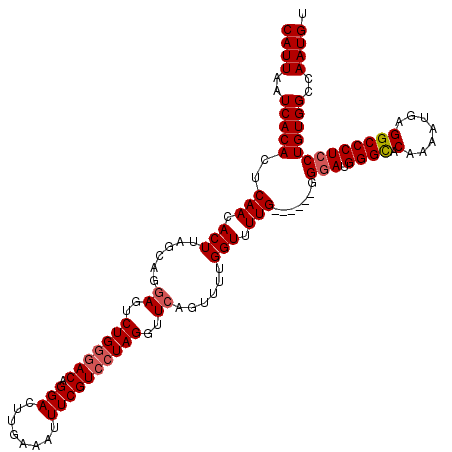
| Location | 5,063,881 – 5,063,995 |
| Length | 114 |
| Max. P | 0.747668 |

| Location | 5,063,881 – 5,063,995 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -29.24 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5063881 114 + 23771897 CAUUAAUCACACUCAACACUUAGCAGGAGUCUGGGACAGGACUUGAAAUUUCGUCCUAGGUUCACAUUUGGUUUUG------GGAAUGGGCACAAAAUGAGGCCCUCCUGUGGCCAAUGU ((((..((((((((((.(((......((..(((((((.(((........))))))))))..))......))).)))------))...((((.(.......)))))...)))))..)))). ( -34.70) >DroSec_CAF1 270401 114 + 1 CAUUAAUCACACUCAACACUUAGCAGGAGUCUGGGACAGGACUUGAAAUUUCGUCCUAGGUUCAGUUUUGGUUUUG------GGGAUGGGCGCAAAAUGAGGCCCUCCUGUGGCCAAUGU ((((..((((...(((.(((.(((..((..(((((((.(((........))))))))))..)).)))..))).)))------((((.((((.(.....)..))))))))))))..)))). ( -36.60) >DroSim_CAF1 291704 114 + 1 CAUUAAUCACACUCAACACUUAGCAGGAGUCUGGGACAGGACUUGAAAUUUCGUCCUAGGUUCAGUUUUGGUUUUG------GGGAGGGGCACAAAAUGAGGCCCUCCUGUGGCCAAUGU ((((..((((...(((.(((.(((..((..(((((((.(((........))))))))))..)).)))..))).)))------(((((((.(.(.....).).)))))))))))..)))). ( -39.50) >DroEre_CAF1 277265 113 + 1 CAUUAAUCACACUCAACACUUAGCAGGAGUCUGCUACAGGACUUGAAAUUUCGUCCUAGGUUCAGUUUUGGUUUUG-------GGAGGGGUUCGAAAUAAGGCCCUCCUGUGGCCAAUGU ..........(((.(((...((((((....)))))).(((((..........)))))..))).))).(((((..((-------((((((.((.......)).))))))))..)))))... ( -35.40) >DroYak_CAF1 281053 120 + 1 CAUUAAUCACACUCAACACUUAGCAGGAGUCUGGGACAGGACUUGAAAUUUCGUCCUAGGUUAAGUUUUGGUUUUGGUUGUGGGGAUGGGCACAGAACCAGGCCCUCCUGUGGCCAAUGU ............((((.(((((((......(((((((.(((........))))))))))))))))).))))..(((((..(((((..((((.(.......))))))))))..)))))... ( -37.30) >consensus CAUUAAUCACACUCAACACUUAGCAGGAGUCUGGGACAGGACUUGAAAUUUCGUCCUAGGUUCAGUUUUGGUUUUG______GGGAUGGGCACAAAAUGAGGCCCUCCUGUGGCCAAUGU ((((..(((((..(((.(((......((..(((((((.(((........))))))))))..))......))).))).......(((.((((.(.......)))))))))))))..)))). (-29.24 = -29.88 + 0.64)


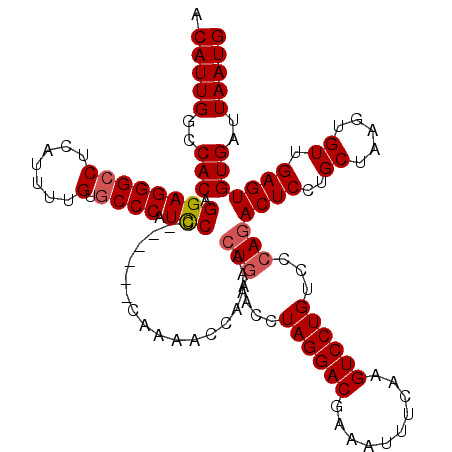
| Location | 5,063,881 – 5,063,995 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -26.14 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5063881 114 - 23771897 ACAUUGGCCACAGGAGGGCCUCAUUUUGUGCCCAUUCC------CAAAACCAAAUGUGAACCUAGGACGAAAUUUCAAGUCCUGUCCCAGACUCCUGCUAAGUGUUGAGUGUGAUUAAUG (((((((.....((((((((.......).))))..)))------.....)).))))).....((((((..........))))))...((.((((..((.....)).)))).))....... ( -27.90) >DroSec_CAF1 270401 114 - 1 ACAUUGGCCACAGGAGGGCCUCAUUUUGCGCCCAUCCC------CAAAACCAAAACUGAACCUAGGACGAAAUUUCAAGUCCUGUCCCAGACUCCUGCUAAGUGUUGAGUGUGAUUAAUG .(((((..(((.((((((((.......).)))).))).------...........(((....((((((..........))))))...)))((((..((.....)).)))))))..))))) ( -30.00) >DroSim_CAF1 291704 114 - 1 ACAUUGGCCACAGGAGGGCCUCAUUUUGUGCCCCUCCC------CAAAACCAAAACUGAACCUAGGACGAAAUUUCAAGUCCUGUCCCAGACUCCUGCUAAGUGUUGAGUGUGAUUAAUG .(((((..(((.((((((.(.(.....).).)))))).------...........(((....((((((..........))))))...)))((((..((.....)).)))))))..))))) ( -30.90) >DroEre_CAF1 277265 113 - 1 ACAUUGGCCACAGGAGGGCCUUAUUUCGAACCCCUCC-------CAAAACCAAAACUGAACCUAGGACGAAAUUUCAAGUCCUGUAGCAGACUCCUGCUAAGUGUUGAGUGUGAUUAAUG .(((((..(((.((((((..((.....))..))))))-------..........(((.((((((((((..........))))).((((((....)))))))).))).))))))..))))) ( -35.60) >DroYak_CAF1 281053 120 - 1 ACAUUGGCCACAGGAGGGCCUGGUUCUGUGCCCAUCCCCACAACCAAAACCAAAACUUAACCUAGGACGAAAUUUCAAGUCCUGUCCCAGACUCCUGCUAAGUGUUGAGUGUGAUUAAUG .(((((..(((.((((((((.......).)))).))).................((((((((((((((..........)))))....(((....)))...)).))))))))))..))))) ( -31.80) >consensus ACAUUGGCCACAGGAGGGCCUCAUUUUGUGCCCAUCCC______CAAAACCAAAACUGAACCUAGGACGAAAUUUCAAGUCCUGUCCCAGACUCCUGCUAAGUGUUGAGUGUGAUUAAUG .(((((..(((.((((((((.......).)))).)))..................(((....((((((..........))))))...)))((((..((.....)).)))))))..))))) (-26.14 = -26.58 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:06 2006