
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,057,542 – 5,057,761 |
| Length | 219 |
| Max. P | 0.999911 |

| Location | 5,057,542 – 5,057,641 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -19.10 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
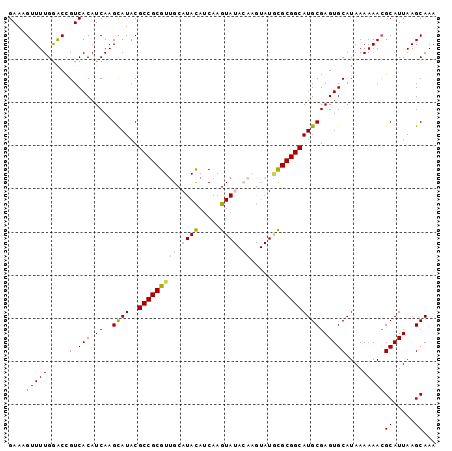
>3L_DroMel_CAF1 5057542 99 + 23771897 GAAAGUUUUGGACUGUCACAUCAAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAA ....(((((....((.(((.((..((((..(((((...((((((.............))))))))))))))))))))))...)))))((.....))... ( -26.42) >DroSec_CAF1 263995 99 + 1 GAAAGUUUUGGACCGUCACAUCAAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAAGGCAUUAAGCAAA ....((((..(.((................(((((...((((((.............)))))))))))((((....))))......)))...))))... ( -23.62) >DroSim_CAF1 285327 99 + 1 GAAAGUUUUGGACCGUCACAUCAAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAA ....(((((.....(.(((.((..((((..(((((...((((((.............)))))))))))))))))))))....)))))((.....))... ( -24.82) >DroEre_CAF1 270776 91 + 1 GAAAGUUUUGGGCCGUCACAUCAAGUAUACGCCGCGUUGCAUAUACCAAGUA--------UAUGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAA ....((((..(((.((.((.....))..)))))(((((((((((((...)))--------))))).((((....))))......)))))...))))... ( -23.80) >DroYak_CAF1 274433 91 + 1 GAAAGUUUUGAACCGUCACAUCAAGUAUACGCCGCGUGGCAUACAUCAAGUA--------UUCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAA ....(((((.....(.(((.((..((((..((((((....((((.....)))--------).))))))))))))))))....)))))((.....))... ( -20.80) >consensus GAAAGUUUUGGACCGUCACAUCAAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAA ....(((((.....(.(((.((..((((..(((((((((.((((.....)))).)).....)))))))))))))))))....)))))((.....))... (-19.10 = -20.02 + 0.92)

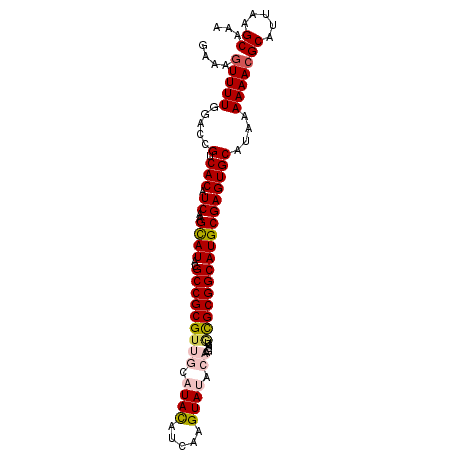
| Location | 5,057,542 – 5,057,641 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -19.82 |
| Energy contribution | -22.26 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
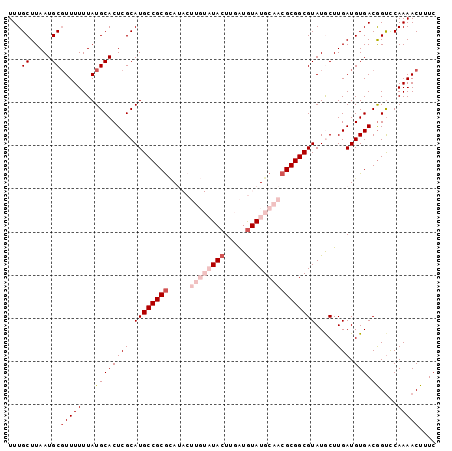
>3L_DroMel_CAF1 5057542 99 - 23771897 UUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUUGAUGUGACAGUCCAAAACUUUC ...((.....))(((((...(((((((((((((((((.....((((((((.....))))))))))))))..))))..)).))).))....))))).... ( -30.00) >DroSec_CAF1 263995 99 - 1 UUUGCUUAAUGCCUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUUGAUGUGACGGUCCAAAACUUUC .(..(((((.((.......((((....))))((((((.....((((((((.....))))))))))))))....)))))).)..)............... ( -26.00) >DroSim_CAF1 285327 99 - 1 UUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUUGAUGUGACGGUCCAAAACUUUC ...((.....))(((((...(((((((((((((((((.....((((((((.....))))))))))))))..))))..)).))).))....))))).... ( -29.30) >DroEre_CAF1 270776 91 - 1 UUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCAUA--------UACUUGGUAUAUGCAACGCGGCGUAUACUUGAUGUGACGGCCCAAAACUUUC ...((.....))(((((..((((....))))((((((((--------(....(((((((((......)))))))))..))))).))))..))))).... ( -28.80) >DroYak_CAF1 274433 91 - 1 UUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGAA--------UACUUGAUGUAUGCCACGCGGCGUAUACUUGAUGUGACGGUUCAAAACUUUC ...((.....))(((((....((..((((((((((((.(--------(((.....))))....))))))(....)...))))))..))..))))).... ( -23.00) >consensus UUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUUGAUGUGACGGUCCAAAACUUUC ...((.....))(((((...((((((((.((((((((.....((((((((.....)))))))))))))))).....))).))).))....))))).... (-19.82 = -22.26 + 2.44)


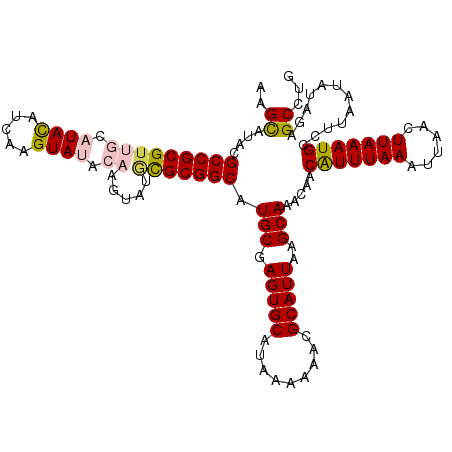
| Location | 5,057,564 – 5,057,681 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057564 117 + 23771897 AAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUG ..((....(((((...((((((.............))))))))))).(((.(((((.........)))))..))).....(((((((......)))))))............))... ( -26.82) >DroSec_CAF1 264017 117 + 1 AAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAAGGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUG ..((....(((((...((((((.............)))))))))))((((....))))....(((((((((((...................))).))))))))........))... ( -28.73) >DroSim_CAF1 285349 117 + 1 AAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUG ..((....(((((...((((((.............))))))))))).(((.(((((.........)))))..))).....(((((((......)))))))............))... ( -26.82) >DroEre_CAF1 270798 109 + 1 AAGUAUACGCCGCGUUGCAUAUACCAAGUA--------UAUGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAAAGAGCCUG ..((((.(((...(((((((((((...)))--------))))))))..))).)))).........((.....))......(((((((......)))))))................. ( -24.90) >DroYak_CAF1 274455 109 + 1 AAGUAUACGCCGCGUGGCAUACAUCAAGUA--------UUCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACGUUUAAAUUAACUUAAAUGCCUUAAUAAAGAGCCUG ((((....((((((....((((.....)))--------).)))))).(((.(((((.........)))))..)))................))))....(((((....))).))... ( -23.20) >consensus AAGCAUACGCCGCGUUGCAUACAUCAAGUAUACAAGUAUGCGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUG ..((....(((((((((.((((.....)))).)).....))))))).(((.(((((.........)))))..))).....(((((((......)))))))............))... (-22.22 = -22.78 + 0.56)


| Location | 5,057,564 – 5,057,681 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -24.64 |
| Energy contribution | -27.44 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057564 117 - 23771897 CAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUU .((((..........((((((((((....))))))))))....((....(((((.....)))))...))((((((((.....((((((((.....))))))))))))))))..)))) ( -34.50) >DroSec_CAF1 264017 117 - 1 CAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCCUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUU .((((..((((..((((((((.((((.(((.........))).))))))))))))...)))).......((((((((.....((((((((.....))))))))))))))))..)))) ( -36.70) >DroSim_CAF1 285349 117 - 1 CAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUU .((((..........((((((((((....))))))))))....((....(((((.....)))))...))((((((((.....((((((((.....))))))))))))))))..)))) ( -34.50) >DroEre_CAF1 270798 109 - 1 CAGGCUCUUUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCAUA--------UACUUGGUAUAUGCAACGCGGCGUAUACUU ...(((....(((((((((((((((....)))))))))......))))))(((((...(((((....)))))..(((((--------(((...))))))))))))))))........ ( -27.70) >DroYak_CAF1 274455 109 - 1 CAGGCUCUUUAUUAAGGCAUUUAAGUUAAUUUAAACGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGAA--------UACUUGAUGUAUGCCACGCGGCGUAUACUU ...((.........(((((..((((((......))).)))..)))))..(((((.....)))))...))((((((((.(--------(((.....))))....))))))))...... ( -28.60) >consensus CAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCGCAUACUUGUAUACUUGAUGUAUGCAACGCGGCGUAUGCUU ...((.....(((((((((((((((....)))))))))......))))))((((.....))))....))((((((((.....((((((((.....))))))))))))))))...... (-24.64 = -27.44 + 2.80)

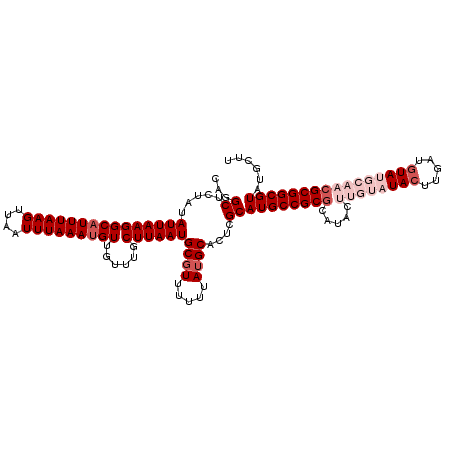
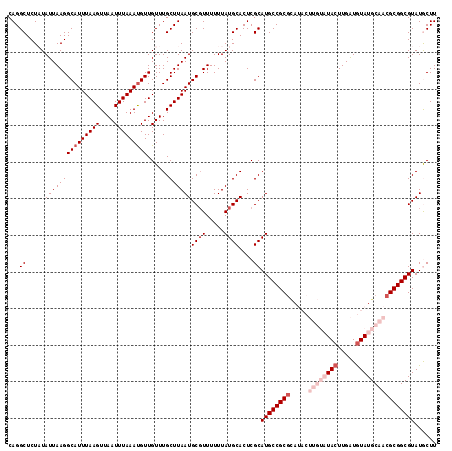
| Location | 5,057,604 – 5,057,721 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 98.46 |
| Mean single sequence MFE | -21.53 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057604 117 + 23771897 CGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUGGCCAAAUACAUUUGACAUAUUUACUUAAUCGCAUUUCGCC .((((.((((((((((.........))))((((.......(((((((......)))))))........((((...((..((((....)))).))..)))))))).)))))).)))). ( -21.30) >DroSec_CAF1 264057 117 + 1 CGCGGCAUGCGAGUGCAUAAAAAAGGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUGGCCAAAUACAUUUGACAUAUUUACUUAAUCGCAUUUCGCC .((((.((((((..........(((((((((((...................))).))))))))....((((...((..((((....)))).))..)))).....)))))).)))). ( -23.81) >DroSim_CAF1 285389 117 + 1 CGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUGGCCAAAUACAUUUGACAUAUUUACUUAAUCGCAUUUCGCC .((((.((((((((((.........))))((((.......(((((((......)))))))........((((...((..((((....)))).))..)))))))).)))))).)))). ( -21.30) >DroEre_CAF1 270830 117 + 1 UGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAAAGAGCCUGGCCAAAUACAUUUGACAUAUUUACUUAAUCGCAUUUCGCC .((((.((((((((((.........))))((((.(((...(((((((......)))))))...............((..((((....)))).))..))).)))).)))))).)))). ( -20.40) >DroYak_CAF1 274487 117 + 1 CGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACGUUUAAAUUAACUUAAAUGCCUUAAUAAAGAGCCUGGCCAAAUACAUUUGACAUAUUUACUUAAUCGCAUUUCGCC .((((.((((((((((.........))))((((.(((....((((((......))))))(((..............))).................))).)))).)))))).)))). ( -20.84) >consensus CGCGGCAUGCGAGUGCAUAAAAAACGCAUUAAGCAAACAACAUUUAAAUUAACUUAAAUGCCUUAAUAUAGAGCCUGGCCAAAUACAUUUGACAUAUUUACUUAAUCGCAUUUCGCC .((((.((((((((((.........))))((((.(((...(((((((......)))))))...............((..((((....)))).))..))).)))).)))))).)))). (-20.70 = -20.54 + -0.16)

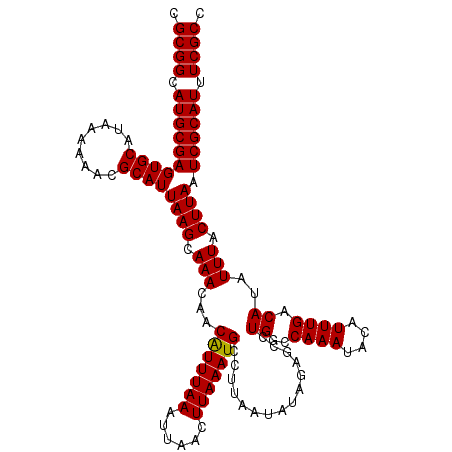

| Location | 5,057,604 – 5,057,721 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 98.46 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -33.40 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057604 117 - 23771897 GGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCG .(((..((((((((((((((((..((((((....))))))...............((((((((((....))))))))))))))))))))(((((.....))))).))))))..))). ( -35.00) >DroSec_CAF1 264057 117 - 1 GGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCCUUUUUUAUGCACUCGCAUGCCGCG .(((..((((((((((((((((..((((((....))))))...............((((((((((....))))))))))))))))))))(((.........))).))))))..))). ( -31.40) >DroSim_CAF1 285389 117 - 1 GGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCG .(((..((((((((((((((((..((((((....))))))...............((((((((((....))))))))))))))))))))(((((.....))))).))))))..))). ( -35.00) >DroEre_CAF1 270830 117 - 1 GGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUUUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCA .(((..((((((((((((((((..((((((....))))))...............((((((((((....))))))))))))))))))))(((((.....))))).))))))..))). ( -35.00) >DroYak_CAF1 274487 117 - 1 GGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUUUAUUAAGGCAUUUAAGUUAAUUUAAACGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCG .(((..(((((((((((((((((.((((((....))))))...((.(((....)))))....................)))))))))))(((((.....))))).))))))..))). ( -32.70) >consensus GGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUGUUUGCUUAAUGCGUUUUUUAUGCACUCGCAUGCCGCG .(((..((((((((((((((((..((((((....))))))...............((((((((((....))))))))))))))))))))(((((.....))))).))))))..))). (-33.40 = -33.80 + 0.40)


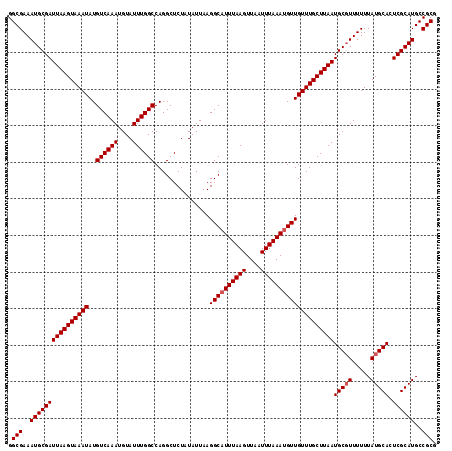
| Location | 5,057,641 – 5,057,761 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057641 120 - 23771897 GAGUUGUUGGCAAAGUAACGUUUGAAACUCAUACGCGUUCGGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUG ((((..(((((.(((((.(((((((....(((((((.....)))...(((......)))..)))))))))))))))).)))))))))........((((((((((....)))))))))). ( -30.80) >DroSec_CAF1 264094 120 - 1 GAGUUGUUGGCAAAGUAACGUUUGAAACUCAUACUCGUUCGGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUG ((((..(((((.(((((.(((((((......((((...(((........)))...))))......)))))))))))).)))))))))........((((((((((....)))))))))). ( -30.90) >DroSim_CAF1 285426 120 - 1 GAGUUGUUGGCAAAGUAACGUUUGAAACUCAUACUCGUUCGGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUG ((((..(((((.(((((.(((((((......((((...(((........)))...))))......)))))))))))).)))))))))........((((((((((....)))))))))). ( -30.90) >DroEre_CAF1 270867 120 - 1 CAGUUGCUGGCAAAGUAACGUUUGAAACUCAAACCCGCUCGGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUUUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUG ..((((((.....))))))(((((.....)))))..((..(((.(((((((((...........)))....)))))).)))..))..........((((((((((....)))))))))). ( -30.60) >DroYak_CAF1 274524 120 - 1 GAGUUGUUGGCAAAGUAACGUUUGAAACCCAUACCCGUACGGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUUUAUUAAGGCAUUUAAGUUAAUUUAAACGUUG ((((..(((((.(((((.(((((((....(((((((....)).)...(((......)))..)))))))))))))))).)))))))))........(((.((((((....)))))).))). ( -23.90) >consensus GAGUUGUUGGCAAAGUAACGUUUGAAACUCAUACCCGUUCGGCGAAAUGCGAUUAAGUAAAUAUGUCAAAUGUAUUUGGCCAGGCUCUAUAUUAAGGCAUUUAAGUUAAUUUAAAUGUUG ((((..(((((.(((((.(((((((......(((....(((........)))....)))......)))))))))))).)))))))))........((((((((((....)))))))))). (-26.76 = -27.40 + 0.64)

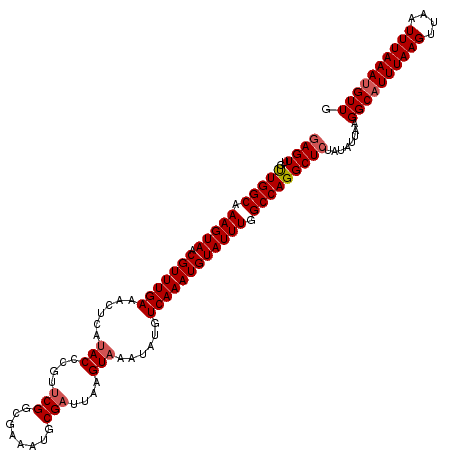
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:50 2006