
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,057,438 – 5,057,538 |
| Length | 100 |
| Max. P | 0.811541 |

| Location | 5,057,438 – 5,057,538 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -10.25 |
| Energy contribution | -10.61 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057438 100 + 23771897 GGGCUUAAAACGCAAUUCUCAAAUUGAAAAUUGUUGUCAUCGUAAAUUGGGAAUAAUUAUUUUUAAUUGUGUGCA-AC-----------------AUGUGGUAAGCGAAU--UGCAUGUG ...........(((.......((((((((((.(((((..(((.....)))..))))).))))))))))...))).-((-----------------(((..((......))--..))))). ( -21.50) >DroPse_CAF1 332180 120 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNAAUUGUUGUCAACGUUAAUUGAGAAUAAUUAUAUUUAAUUGCAUGGCUGCAAGAGCAUGCAGCCGGUAUGCGGCAGCCAAAUGUGGCAUGUG ............................(((((((.((((......)))).)))))))........(..(((((((((....((((((.....)))))).)))))))..))..)...... ( -30.60) >DroSec_CAF1 263891 100 + 1 GGGCUUAAAACGCAAUUCUCAAAUUGAAAAUUGUUGUCAUCGUAAAUUGGGAAUAAUUAUUUUUAAUUGUGUGCC-AC-----------------AUGUGGUAAGCGAAU--UGCAUGUG ...........(((((((...((((((((((.(((((..(((.....)))..))))).))))))))))((.((((-(.-----------------...))))).))))))--)))..... ( -26.80) >DroSim_CAF1 285224 99 + 1 GGGCUUA-AACGCAAUUCUCAAAUUGAAAAUUGUUGUCAUCGUAAAUUGGGAAUAAUUAUUUUUAAUUGUGUGCC-AC-----------------AUGUGGUAAGCGAAU--UGCAUGUG .......-...(((((((...((((((((((.(((((..(((.....)))..))))).))))))))))((.((((-(.-----------------...))))).))))))--)))..... ( -26.80) >DroEre_CAF1 270677 95 + 1 -----UAAAACGCAAUUCUCAAAUUGAAAAUUGUUGUCAUCGUAAAUUGGGAAUAAUUAUUUUUAAUUGUAUGCC-GC-----------------GUGUGGUAAGCGAAU--UGCAUGUG -----......(((((((...((((((((((.(((((..(((.....)))..))))).))))))))))((.((((-(.-----------------...))))).))))))--)))..... ( -26.10) >DroYak_CAF1 274327 102 + 1 GGGCUUAAAACGCAAUUCUCAAAUUGAAAAUUGUUGUCAUCGUAAAUUGGGAAUAAUUAUUUUUAAUUGUAAGCC-GCAU---------------GUGUGGUAAGCGAAU--UGCAUGUG ...........(((((((...((((((((((.(((((..(((.....)))..))))).))))))))))((..(((-((..---------------..)))))..))))))--)))..... ( -26.80) >consensus GGGCUUAAAACGCAAUUCUCAAAUUGAAAAUUGUUGUCAUCGUAAAUUGGGAAUAAUUAUUUUUAAUUGUAUGCC_AC_________________AUGUGGUAAGCGAAU__UGCAUGUG ...........(((..............(((((((.(((........))).)))))))........((((.((((.((...................)))))).))))....)))..... (-10.25 = -10.61 + 0.36)

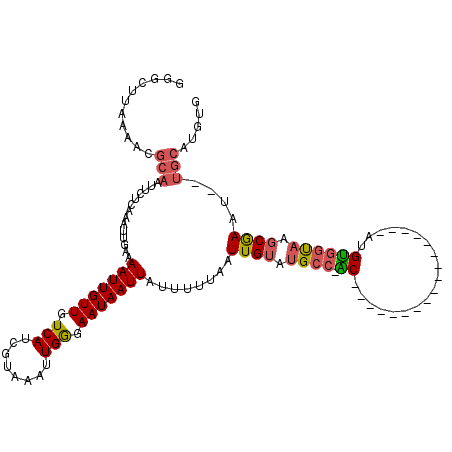

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:44 2006