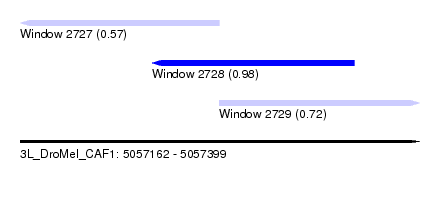
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,057,162 – 5,057,399 |
| Length | 237 |
| Max. P | 0.980488 |

| Location | 5,057,162 – 5,057,280 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.97 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057162 118 - 23771897 UUGCUAAGCAAUUAAAAAAUAUAGAAAAAAAAAAUGGCAUUGAUUGCAGCUGCCACCGCACACCGACCAUGUGGCAC--AUAGUUCUAUAGUUCCAAAGCCGCACGUGUGCUUUUUUAUC ((((...)))).........((((((((......((((((((....))).)))))..((((((......((((((..--...................)))))).)))))))))))))). ( -25.60) >DroSec_CAF1 263639 94 - 1 UUGCUAAGCAAUUAAAAAAUAUAGAAAAAA---ACUG---------------------CACACCGACCAUGUGGCAC--AUAGUUCUAUAGUUCCAAAGCCGCACGUGUGCUUUUUUAUC ((((...))))............((.((((---(..(---------------------(((((......((((((..--...................)))))).)))))).))))).)) ( -16.70) >DroSim_CAF1 284952 115 - 1 UUGCUAAGCAAUUAAAAAAUAUAGAAAAAA---ACUGCAUUGAUUGCAGCUGCCACCGCACACCGACCAUGUGGCAC--AUAGUUCUAUAGUUCCAAAGCCGCACGUGUGCUUUUUUAUC ((((...)))).........((((((((..---.(((((.....)))))........((((((......((((((..--...................)))))).)))))))))))))). ( -24.50) >DroEre_CAF1 270412 115 - 1 UUGCUAAGCAAUUAAAAAAUAUAGAAAAAA---ACUGCAUUGAUUGCAGCUGCCACCGCACACCUACCAUGUGGCAC--AUAGUUCUAUAGUUCCAAAGCCGCACGUGUGCUUUUUUAUC ((((...)))).........((((((((..---.(((((.....)))))........((((((......((((((..--...................)))))).)))))))))))))). ( -24.50) >DroYak_CAF1 274048 120 - 1 UUGCUAAGCAAUUAAAAAAUAUAGAAAAAAAAAGCUGCAUUGAUUGCAGCUGCCACCGCACACCGACCAUGUGGCACACACAGUUCUAUAGUUCCAAAGCCGCACGUGUGCUUUUUUAUC ((((...)))).........((((((((....(((((((.....)))))))......((((((......((((((.......................)))))).)))))))))))))). ( -29.50) >consensus UUGCUAAGCAAUUAAAAAAUAUAGAAAAAA___ACUGCAUUGAUUGCAGCUGCCACCGCACACCGACCAUGUGGCAC__AUAGUUCUAUAGUUCCAAAGCCGCACGUGUGCUUUUUUAUC ((((...)))).........((((((((......(((((.....)))))........((((((......((((((.......................)))))).)))))))))))))). (-18.88 = -19.86 + 0.98)


| Location | 5,057,240 – 5,057,360 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.62 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057240 120 - 23771897 AUUCAGUUCGGAACCGAUAGCGGAUGCAGUUGGCAAAGGAAGCCUUGACCAUCCGCUCCAAUUGUUCCGACGCGCUUCCUUUGCUAAGCAAUUAAAAAAUAUAGAAAAAAAAAAUGGCAU .(((.(((((((((.(((((((((((.....(((.......))).....))))))))...)))))))))).))((((........))))..............))).............. ( -31.90) >DroSec_CAF1 263699 114 - 1 AUUCAGUUCGGAACCGAUAGCGGAUGCAGCUGGCAAAGGAAGCCCUGACCAUCCGCUCCAAUUGUUCCGACGCGCUUCCUUUGCUAAGCAAUUAAAAAAUAUAGAAAAAA---ACUG--- .(((.(((((((((.((((((((((((((..(((.......))))))..))))))))...)))))))))).))((((........))))..............)))....---....--- ( -32.30) >DroSim_CAF1 285030 117 - 1 AUUCAGUUCGGAACCGAUAGCGGAUGCAGCUGGCAAAGGAAGCCCUGACCAUCCGCUCCAAUUGUUCCGACGCGCUUCCUUUGCUAAGCAAUUAAAAAAUAUAGAAAAAA---ACUGCAU .(((.(((((((((.((((((((((((((..(((.......))))))..))))))))...)))))))))).))((((........))))..............)))....---....... ( -32.30) >DroEre_CAF1 270490 117 - 1 GCUCAGUUCGGAGCCGAUAGCGGAUGCAGCUGCCAAAGGAAGCCCUGACCAUCCGCUCCAAUUGUUCCGACGCGCUUCCUUUGCUAAGCAAUUAAAAAAUAUAGAAAAAA---ACUGCAU ((...(((((((((.((((((((((((((..((........)).)))..))))))))...)))))))))).))((((........)))).....................---...)).. ( -29.90) >DroYak_CAF1 274128 120 - 1 AUUCAGUUCGCCACCGAUAGCGGAUGCAGCUGCCAAAGGAAGCCCUGACCAUCCGCUCCAAUUGUUCCGACGCGCUUCCUUUGCUAAGCAAUUAAAAAAUAUAGAAAAAAAAAGCUGCAU .....((((((........))))))((((((((.(((((((((...(......)((((..........)).)))))))))))))(((....)))..................)))))).. ( -29.90) >consensus AUUCAGUUCGGAACCGAUAGCGGAUGCAGCUGGCAAAGGAAGCCCUGACCAUCCGCUCCAAUUGUUCCGACGCGCUUCCUUUGCUAAGCAAUUAAAAAAUAUAGAAAAAA___ACUGCAU .(((.(((((((((.((((((((((((((..(((.......))))))..))))))))...)))))))))).))((((........))))..............))).............. (-26.82 = -27.50 + 0.68)

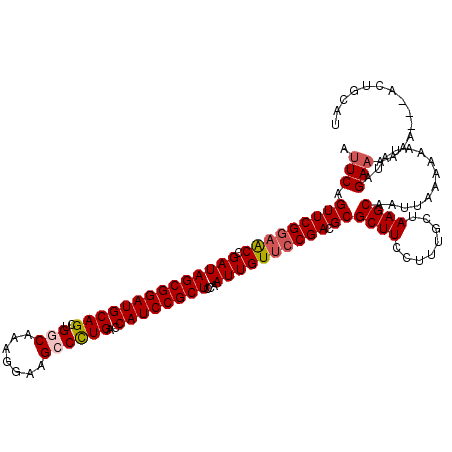
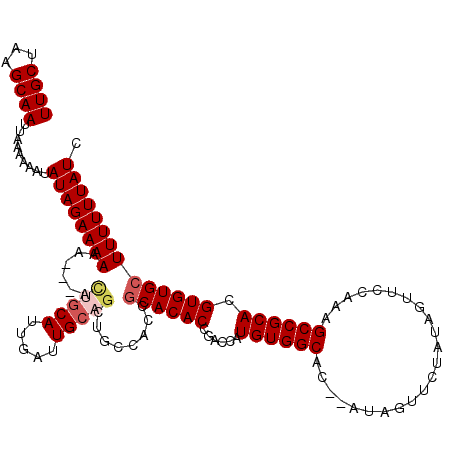
| Location | 5,057,280 – 5,057,399 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -41.21 |
| Consensus MFE | -35.56 |
| Energy contribution | -35.72 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5057280 119 + 23771897 AGGAAGCGCGUCGGAACAAUUGGAGCGGAUGGUCAAGGCUUCCUUUGCCAACUGCAUCCGCUAUCGGUUCCGAACUGAAUUGAAUAGCGACCAAAGACGCCGUAAUUGACGUUGAUCCA- .(((...(((((.........(((((((.((((.((((...)))).)))).)))).)))(((((((((((......)))))).))))).......)))))(((.....)))....))).- ( -41.70) >DroSec_CAF1 263733 119 + 1 AGGAAGCGCGUCGGAACAAUUGGAGCGGAUGGUCAGGGCUUCCUUUGCCAGCUGCAUCCGCUAUCGGUUCCGAACUGAAUUGAAAAGCGACCAAAGACGCCGUAAUUGACGUUGAUCCA- .(((...((((((((((......((((((((..((((((.......)))..)))))))))))....)))))......(((((....(((........)))..))))))))))...))).- ( -41.00) >DroSim_CAF1 285067 119 + 1 AGGAAGCGCGUCGGAACAAUUGGAGCGGAUGGUCAGGGCUUCCUUUGCCAGCUGCAUCCGCUAUCGGUUCCGAACUGAAUUGAAAAGCGACCAAAGACGCCGUAAUUGACGUUGAUCCA- .(((...((((((((((......((((((((..((((((.......)))..)))))))))))....)))))......(((((....(((........)))..))))))))))...))).- ( -41.00) >DroEre_CAF1 270527 120 + 1 AGGAAGCGCGUCGGAACAAUUGGAGCGGAUGGUCAGGGCUUCCUUUGGCAGCUGCAUCCGCUAUCGGCUCCGAACUGAGCUGAAAUGCGACCAAAGACGCCGUAAUUGACGUUGAUCCAG .(((((((((.(((.....((((((((((((..(((.(((......)))..))))))))))).(((((((......))))))).......)))).....)))....)).))))..))).. ( -43.30) >DroYak_CAF1 274168 119 + 1 AGGAAGCGCGUCGGAACAAUUGGAGCGGAUGGUCAGGGCUUCCUUUGGCAGCUGCAUCCGCUAUCGGUGGCGAACUGAAUUGAAAAGCGACCAAAGACGCCGUAAUUGACGUUGAUCCA- .(((...(((((.........((((((((((..(((.(((......)))..))))))))))).))(((.((...............)).)))...)))))(((.....)))....))).- ( -39.06) >consensus AGGAAGCGCGUCGGAACAAUUGGAGCGGAUGGUCAGGGCUUCCUUUGCCAGCUGCAUCCGCUAUCGGUUCCGAACUGAAUUGAAAAGCGACCAAAGACGCCGUAAUUGACGUUGAUCCA_ .(((...(((((.........(((((((.((((.((((...)))).)))).)))).)))((..(((((((......)))))))...)).......)))))(((.....)))....))).. (-35.56 = -35.72 + 0.16)

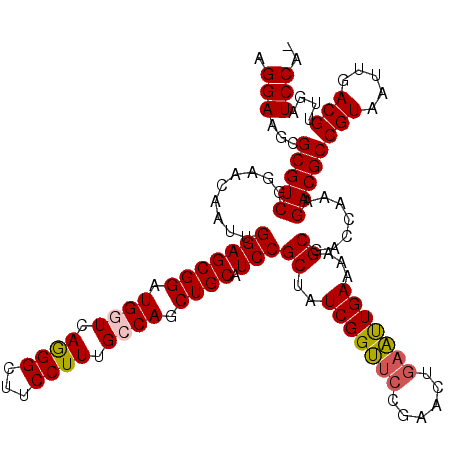
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:43 2006