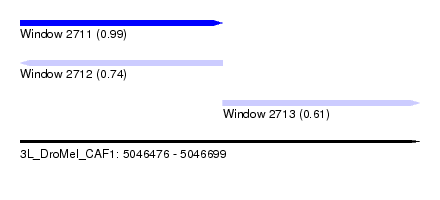
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,046,476 – 5,046,699 |
| Length | 223 |
| Max. P | 0.992949 |

| Location | 5,046,476 – 5,046,589 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5046476 113 + 23771897 ACCCUAAUCAGAGCUCGGCGAAAUGUAAUUUUAAUUUCGCAGCAAUUUGCAAUGCAGAUUGCGUUU-UUUUUUUUUUUUUUUUGUCAUUACAAAAUUUGAGCAUUAUGCAUACA ............((((((((((((.........))))))).(((((((((...)))))))))....-............((((((....))))))..)))))............ ( -27.30) >DroSec_CAF1 252997 102 + 1 GCCCUAAUCAGAGCUCGGCGAAAUGUAAUUUUAAUUUCGGAGCAAUUUGCAAUGCAGAUUGCGUUU-U-----------UUCUGGCAUUACAAAAUUUGAGCAUUAUGCAUACA (((......(((((....((((((.........))))))..(((((((((...)))))))))))))-)-----------....)))..............((.....))..... ( -22.90) >DroSim_CAF1 274285 102 + 1 GCCCUAAUCAGAGCUCGGCGAAAUGUAAUUUUAAUUUCGCAGCAAUUUGCAAUGCAGAUUGCGUUU-U-----------UUCUGGCAUUACAAAAUUUGAGCAUUAUGCAUACA (((......(((((...(((((((.........))))))).(((((((((...)))))))))))))-)-----------....)))..............((.....))..... ( -26.50) >DroEre_CAF1 259788 102 + 1 GCACUAAUCAGAGCUCGGUGAAAUAUAAUUUUAAUUUCGCAGCAAUUUGCAAUGCAGAUUGCGUUU-U-----------UUCUGGCAUUACAAAAUUUGAGCAUUAUGCAUACA (((.((((....((((((((((((.........))))))).(((((((((...)))))))))....-.-----------..................))))))))))))..... ( -25.00) >DroYak_CAF1 262931 103 + 1 GCACUAAUCAGAGCUCGGCGAAAUGUAAUUUUAAUUUCACAGCAAUUUGCCAUGCAGAUCUCGUUUUU-----------UUCUGUCAUUACCAAAUUUGAGCAUUAUGCAUACA (((.((((....((((((((((.(((...............))).))))))(((((((..........-----------.)))).)))..........)))))))))))..... ( -21.26) >consensus GCCCUAAUCAGAGCUCGGCGAAAUGUAAUUUUAAUUUCGCAGCAAUUUGCAAUGCAGAUUGCGUUU_U___________UUCUGGCAUUACAAAAUUUGAGCAUUAUGCAUACA ............((((((((((((.........))))))).(((((((((...)))))))))...................................)))))............ (-21.20 = -21.88 + 0.68)

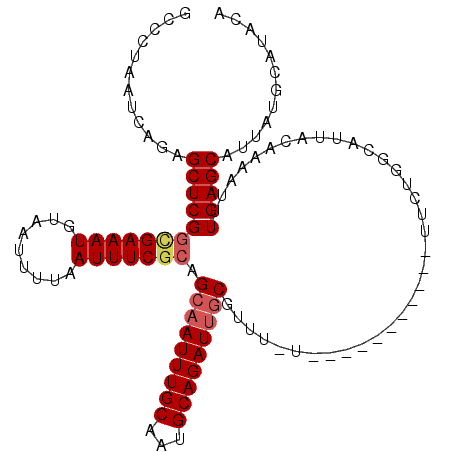
| Location | 5,046,476 – 5,046,589 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -17.95 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5046476 113 - 23771897 UGUAUGCAUAAUGCUCAAAUUUUGUAAUGACAAAAAAAAAAAAAAAA-AAACGCAAUCUGCAUUGCAAAUUGCUGCGAAAUUAAAAUUACAUUUCGCCGAGCUCUGAUUAGGGU ............((((...((((((....))))))............-....(((((.(((...))).))))).(((((((.........))))))).))))............ ( -21.40) >DroSec_CAF1 252997 102 - 1 UGUAUGCAUAAUGCUCAAAUUUUGUAAUGCCAGAA-----------A-AAACGCAAUCUGCAUUGCAAAUUGCUCCGAAAUUAAAAUUACAUUUCGCCGAGCUCUGAUUAGGGC ............((((....((((((((((.(((.-----------.-........)))))))))))))..((((((((((.........))))))..))))........)))) ( -23.80) >DroSim_CAF1 274285 102 - 1 UGUAUGCAUAAUGCUCAAAUUUUGUAAUGCCAGAA-----------A-AAACGCAAUCUGCAUUGCAAAUUGCUGCGAAAUUAAAAUUACAUUUCGCCGAGCUCUGAUUAGGGC ............((((....((((((((((.(((.-----------.-........)))))))))))))..((((((((((.........)))))))..)))........)))) ( -24.30) >DroEre_CAF1 259788 102 - 1 UGUAUGCAUAAUGCUCAAAUUUUGUAAUGCCAGAA-----------A-AAACGCAAUCUGCAUUGCAAAUUGCUGCGAAAUUAAAAUUAUAUUUCACCGAGCUCUGAUUAGUGC .((((.......((((....((((((((((.(((.-----------.-........))))))))))))).......(((((.........)))))...))))........)))) ( -21.36) >DroYak_CAF1 262931 103 - 1 UGUAUGCAUAAUGCUCAAAUUUGGUAAUGACAGAA-----------AAAAACGAGAUCUGCAUGGCAAAUUGCUGUGAAAUUAAAAUUACAUUUCGCCGAGCUCUGAUUAGUGC .((((.......((((.((((((.((.((.((((.-----------..........)))))))).))))))...(((((((.........))))))).))))........)))) ( -19.26) >consensus UGUAUGCAUAAUGCUCAAAUUUUGUAAUGCCAGAA___________A_AAACGCAAUCUGCAUUGCAAAUUGCUGCGAAAUUAAAAUUACAUUUCGCCGAGCUCUGAUUAGGGC ............((((....(((((((((.((((......................))))))))))))).....(((((((.........))))))).))))............ (-17.95 = -18.83 + 0.88)


| Location | 5,046,589 – 5,046,699 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.37 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5046589 110 + 23771897 GUAUUCAUAUGGAUCAUCACAACGGCUGUCAUAAGUGACGUGAUUGAUUGAUGAUGUCACUUAAAGCGACCAACGAAAUAACAGC---AGCAGCAGUUACAACAGAAAU-GUUC (.((((....))))).....(((.(((((..((((((((((.(((....))).))))))))))..((................))---.))))).)))..((((....)-))). ( -26.59) >DroSec_CAF1 253099 110 + 1 GUAUUCAUAUGGAUCAUCACAACGGCUGUCAUAAGUGACGUGAUUGAUUGAUGAUGUCACUUAAAGCGACCAACGAAAUAACAGC---AGCAGCAGUUACAACAGAAAU-GUUC (.((((....))))).....(((.(((((..((((((((((.(((....))).))))))))))..((................))---.))))).)))..((((....)-))). ( -26.59) >DroSim_CAF1 274387 110 + 1 GUAUUCAUAUGGAUCAUCACAACGGCUGUCAUAAGUGACGUGAUUGAUUGAUGAUGUCACUUAAAGCGACCAACGAAAUAACAGC---AGCAGCAGUUACAACAGAAAU-GUUC (.((((....))))).....(((.(((((..((((((((((.(((....))).))))))))))..((................))---.))))).)))..((((....)-))). ( -26.59) >DroEre_CAF1 259890 110 + 1 GUAUUCAUAUAGAGCACCACGCUGGCUGUCAAAAGUGACGUGAUUGAUUGAUGAUGUCACUUAAAGCGACCAACGAAAUAACAGC---AGCGGCAGUUACAAAAAAAAU-GUUC ...........(((((...((.(((.(((...(((((((((.(((....))).)))))))))...))).))).))...((((.((---....)).)))).........)-)))) ( -26.60) >DroYak_CAF1 263034 114 + 1 GUAUUCAUAUAGAGCACCACGACGGCUGUCAUAAGUGACGUGAUUGAUUGAUGAUGUCACUUAAAGCGACCAACGAAAUAUCAGCAGCAGCAGCAGUUACAACAAAAACGGUUC ...............(((.((..((.(((..((((((((((.(((....))).))))))))))..))).))..)).......(((.((....)).)))...........))).. ( -25.70) >consensus GUAUUCAUAUGGAUCAUCACAACGGCUGUCAUAAGUGACGUGAUUGAUUGAUGAUGUCACUUAAAGCGACCAACGAAAUAACAGC___AGCAGCAGUUACAACAGAAAU_GUUC ....................(((.(((((..((((((((((.(((....))).))))))))))...((.....))..............))))).)))................ (-21.16 = -21.00 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:27 2006