
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,045,050 – 5,045,267 |
| Length | 217 |
| Max. P | 0.965593 |

| Location | 5,045,050 – 5,045,160 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -22.31 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5045050 110 - 23771897 GCAAAGCG-----UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGGAGGCAAAUAAACGAGCCGGCAGC----AGAAUCAGCAUCA ((((((((-----(........))...((((....))))))))).(((...(((..........((((.......)))).(((..........))).)))))----)......)).... ( -21.20) >DroSec_CAF1 251571 110 - 1 GCAAAGCG-----UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGGAGGCAAAUAAACGAGCCGGCAGC----AGAAUCAGCAUCA ((((((((-----(........))...((((....))))))))).(((...(((..........((((.......)))).(((..........))).)))))----)......)).... ( -21.20) >DroSim_CAF1 272472 110 - 1 GCAAAGCG-----UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGGAGGCAAAUAAACGAGCCGGCAGC----AGAAUCAGCAUCA ((((((((-----(........))...((((....))))))))).(((...(((..........((((.......)))).(((..........))).)))))----)......)).... ( -21.20) >DroEre_CAF1 255000 115 - 1 GGAAAGCGUAGAACAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGGAGGCAAAUAAGCGAGCUGGCAGC----AGAAUCAGCAUCA .....((...............))...((........(((((((((((....))))))).....((((.......))))..........))))(((((....----....))))).)). ( -22.96) >DroYak_CAF1 261394 114 - 1 GAAAAGCG-----CAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGGAGGCAAAUAAGCGAGUCGGCAGCAGGCAGAAUCAGCAGCA .....(((-----(........((...((((....))))(((((((((....))))))......((((.......)))).)))......))((.((.((.....)).)).)).)).)). ( -25.00) >consensus GCAAAGCG_____UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGGAGGCAAAUAAACGAGCCGGCAGC____AGAAUCAGCAUCA ......................((...((((....))))(((((((((....))))))).....((((.......)))).(((..........))).))..............)).... (-18.96 = -18.64 + -0.32)


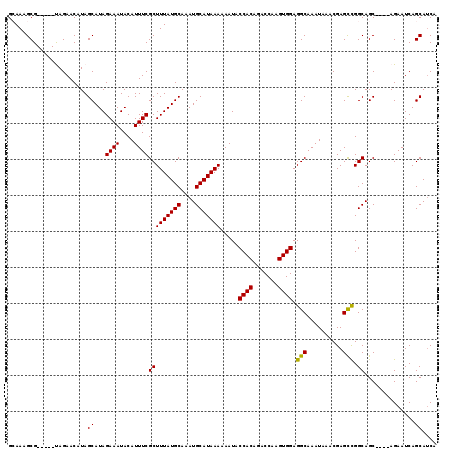
| Location | 5,045,086 – 5,045,191 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -19.50 |
| Consensus MFE | -17.54 |
| Energy contribution | -17.26 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5045086 105 + 23771897 CCACUUGGUCUGUGGUAUUUUUUAUGCAUUUGCAUAAAGCGAAAUGUAUUUCUAUGCUAUGUUCUA-----CGCUUUGCCAACAUUUUCACUACUGUUUUUUCUGGCGCU .....(((..((((((((.....((((((((((.....)).))))))))....))))))))..)))-----......((((.......((....)).......))))... ( -19.94) >DroSec_CAF1 251607 105 + 1 CCACUUGGUCUGUGGUAUUUUUUAUGCAUUUGCAUAAAGCGAAAUGUAUUUCUAUGCUAUGUUCUA-----CGCUUUGCCAACAUUUUCACUACUGUUUUUUCUGGCGCU .....(((..((((((((.....((((((((((.....)).))))))))....))))))))..)))-----......((((.......((....)).......))))... ( -19.94) >DroSim_CAF1 272508 105 + 1 CCACUUGGUCUGUGGUAUUUUUUAUGCAUUUGCAUAAAGCGAAAUGUAUUUCUAUGCUAUGUUCUA-----CGCUUUGCCAACAUUUUCACUACUGUUUUUUCUGGCGCU .....(((..((((((((.....((((((((((.....)).))))))))....))))))))..)))-----......((((.......((....)).......))))... ( -19.94) >DroEre_CAF1 255036 110 + 1 CCACUUGGUCUGUGGUAUUUUUUAUGCAUUUGCAUAAAGCGAAAUGUAUUUCUAUGCUAUGUUCUGUUCUACGCUUUCCCAACAUUUUCACUACUGUUUUUUCUGGCGCU ......((..((((((((.....((((((((((.....)).))))))))....))))))))..))......((((.....((((..........))))......)))).. ( -18.80) >DroYak_CAF1 261434 105 + 1 CCACUUGGUCUGUGGUAUUUUUUAUGCAUUUGCAUAAAGCGAAAUGUAUUUCUAUGCUAUGUUCUG-----CGCUUUUCCAACAUUUUCACUACUGUUUUUUCUGGGUCU ..(((..(.(.(((((....(((((((....)))))))(((((..((((....))))....))).)-----).................))))).)......)..))).. ( -18.90) >consensus CCACUUGGUCUGUGGUAUUUUUUAUGCAUUUGCAUAAAGCGAAAUGUAUUUCUAUGCUAUGUUCUA_____CGCUUUGCCAACAUUUUCACUACUGUUUUUUCUGGCGCU .....(((..((((((((.....((((((((((.....)).))))))))....))))))))..)))...........((((.......((....)).......))))... (-17.54 = -17.26 + -0.28)


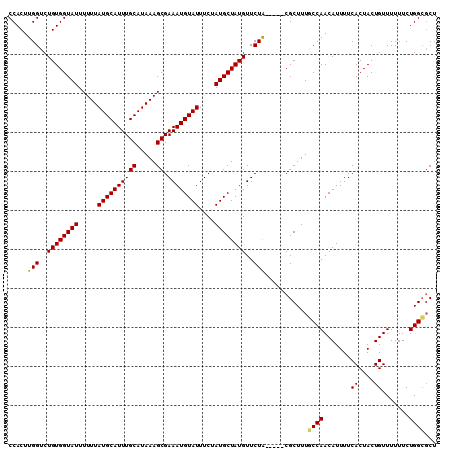
| Location | 5,045,086 – 5,045,191 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5045086 105 - 23771897 AGCGCCAGAAAAAACAGUAGUGAAAAUGUUGGCAAAGCG-----UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGG .(((((((.....((....)).......)))))...((.-----.........))...((((....))))))(((((((....))))))).....((((.......)))) ( -20.20) >DroSec_CAF1 251607 105 - 1 AGCGCCAGAAAAAACAGUAGUGAAAAUGUUGGCAAAGCG-----UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGG .(((((((.....((....)).......)))))...((.-----.........))...((((....))))))(((((((....))))))).....((((.......)))) ( -20.20) >DroSim_CAF1 272508 105 - 1 AGCGCCAGAAAAAACAGUAGUGAAAAUGUUGGCAAAGCG-----UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGG .(((((((.....((....)).......)))))...((.-----.........))...((((....))))))(((((((....))))))).....((((.......)))) ( -20.20) >DroEre_CAF1 255036 110 - 1 AGCGCCAGAAAAAACAGUAGUGAAAAUGUUGGGAAAGCGUAGAACAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGG ...................(((((((((((.....)))))................((....))..))))))(((((((....))))))).....((((.......)))) ( -16.80) >DroYak_CAF1 261434 105 - 1 AGACCCAGAAAAAACAGUAGUGAAAAUGUUGGAAAAGCG-----CAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGG ...................((((((..(((.....)))(-----(........))...........))))))(((((((....))))))).....((((.......)))) ( -17.40) >consensus AGCGCCAGAAAAAACAGUAGUGAAAAUGUUGGCAAAGCG_____UAGAACAUAGCAUAGAAAUACAUUUCGCUUUAUGCAAAUGCAUAAAAAAUACCACAGACCAAGUGG ...................((((((.(((.......((...............))........)))))))))(((((((....))))))).....((((.......)))) (-16.32 = -16.32 + 0.00)


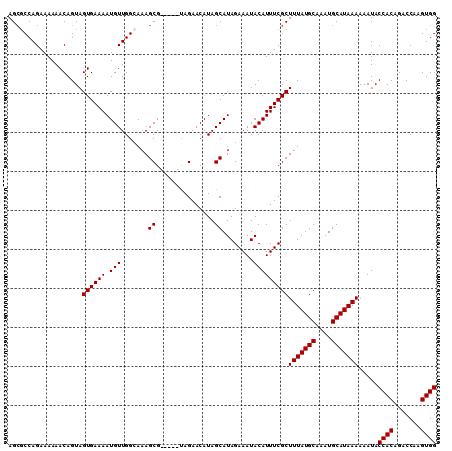
| Location | 5,045,160 – 5,045,267 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 98.88 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -26.81 |
| Energy contribution | -27.41 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5045160 107 - 23771897 GGUGCCCAUCGCCGGGCGCUCAUUAAAAUGCAACUGUGUGCGGUAAUGGCCAAAAAGGGAAACGUGCAAUAAAAGCAGCGCCAGAAAAAACAGUAGUGAAAAUGUUG .((((((......))))))((((((........(((.(((((((....)))......(....).(((.......)))))))))).........))))))........ ( -30.03) >DroSec_CAF1 251681 107 - 1 GGUGCCCAUCGCCGGGCGCUCAUUAAAAUGCAACUGUGUGCGGUAAUGGCCAAAAAGGGAAACGUGCAAUAAAAGCAGCGCCAGAAAAAACAGUAGUGAAAAUGUUG .((((((......))))))((((((........(((.(((((((....)))......(....).(((.......)))))))))).........))))))........ ( -30.03) >DroSim_CAF1 272582 107 - 1 GGUGCCCAUCGCCGGGCGCUCAUUAAAAUGCAACUGUGUGCGGUAAUGGCCAAAAAGGGAAACGUGCAAUAAAAGCAGCGCCAGAAAAAACAGUAGUGAAAAUGUUG .((((((......))))))((((((........(((.(((((((....)))......(....).(((.......)))))))))).........))))))........ ( -30.03) >DroEre_CAF1 255115 107 - 1 GGUGCCCAUCGCCGGGCGCUCAUUAAAAUGCAACUGUGUGCGGUAAUGGGCAAAAAGGGAAACGUGCAAUAAAAGCAGCGCCAGAAAAAACAGUAGUGAAAAUGUUG (((((((((..(((.((((................)))).)))..))))))......(....).(((.......)))..)))......((((..........)))). ( -30.89) >DroYak_CAF1 261508 107 - 1 GGUGCCCAUCGCCGGGCGCUCAUUAAAAUGCAACUGUGUGCGGUAAUGGCCAAAAAGGGAAACGUGCAAUAAAAGCAGACCCAGAAAAAACAGUAGUGAAAAUGUUG (((((((......))))))).........((.(((((....(((....))).....(((...(.(((.......)))).))).......))))).)).......... ( -26.00) >consensus GGUGCCCAUCGCCGGGCGCUCAUUAAAAUGCAACUGUGUGCGGUAAUGGCCAAAAAGGGAAACGUGCAAUAAAAGCAGCGCCAGAAAAAACAGUAGUGAAAAUGUUG .((((((......))))))((((((........(((.(((((((....)))......(....).(((.......)))))))))).........))))))........ (-26.81 = -27.41 + 0.60)

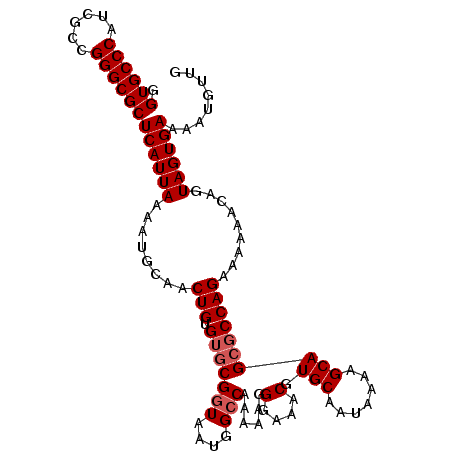

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:23 2006