
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,042,672 – 5,042,812 |
| Length | 140 |
| Max. P | 0.659570 |

| Location | 5,042,672 – 5,042,772 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.51 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -8.58 |
| Energy contribution | -8.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5042672 100 + 23771897 UCUCCU--UCUAUCCCUCGCUCUCUUCGACGGGCACA-CAGUGCGUAUGAUUAAUUUCCCUAAUUU---------UGGGUAUGUGUG--------CCUUCUUGAGAUCACAAAUUCCCAU ......--..........(.((((...((.(((((((-((.(((.((.(((((.......))))).---------)).)))))))))--------)))))..)))).)............ ( -25.20) >DroPse_CAF1 318317 108 + 1 CCCCCCUUCCCUUCUUUGGCACUCUUUUGGGGGCACUCCAGUGCGUAUGAUUAAUUUGCUUAAUUU---------UGUGUAUGAGGCACAGAAAUCCCACCCGAAA---CGCCACCGCUU ................((((....(((((((.((((....))))...................(((---------(((((.....))))))))......)))))))---.))))...... ( -26.10) >DroSec_CAF1 249159 102 + 1 UCUCCUUUUCUGUCCCUCGCUCUCUUCGAGGGGCACA-CAGUGCGUAUGAUUAAUUUCCUUAAUUU---------UGGGUAUGUGUG--------CCUACUUGAGAAGACGAAUUCCCAU ................(((.(((..((((((((((((-((.(((.((.((((((.....)))))).---------)).)))))))))--------))).)))))..))))))........ ( -32.90) >DroSim_CAF1 270020 102 + 1 UCUCCUUUUCUCUCCCUCGCUCUCUUCGAGGGGCACA-CAGUGCGUAUGAUUAAUUUCCUUAAUUU---------UGGGUAUGUGUG--------CCUACUUGAGAAGACGAAUUCCCAU ..((.(((((((..(((((.......)))))((((((-((.(((.((.((((((.....)))))).---------)).)))))))))--------)).....))))))).))........ ( -34.30) >DroEre_CAF1 252603 98 + 1 UCCACA--UCUGUCCC-CGCUCUCUCCUUCG-GCACA-CAGUGCGUAUGAUUAAUUUCCUUAAUUU---------UGGGUAUGUGUG--------CCUUCUUGAGAAGACGAAUUCCCAU ......--........-((.((((((....(-(((((-((.(((.((.((((((.....)))))).---------)).)))))))))--------)).....))).)))))......... ( -25.60) >DroYak_CAF1 259006 109 + 1 UCGACU--UCUGUCCCUCGCUCUCUUCAACGGGCACA-CAGUGCGUAUGAUUAAUUUCCUUAAUUUUUGGGUUUGUGGGUAUGUGUG--------CCUACUUGCGAAGACGAAUUUCCAU ..(((.--...)))..(((.(((...(((.(((((((-((.(((.((..(.......(((........))).)..)).)))))))))--------)))..)))...))))))........ ( -27.60) >consensus UCUCCU__UCUGUCCCUCGCUCUCUUCGACGGGCACA_CAGUGCGUAUGAUUAAUUUCCUUAAUUU_________UGGGUAUGUGUG________CCUACUUGAGAAGACGAAUUCCCAU ..................((((..........((((....))))....((((((.....))))))...........))))........................................ ( -8.58 = -8.47 + -0.11)


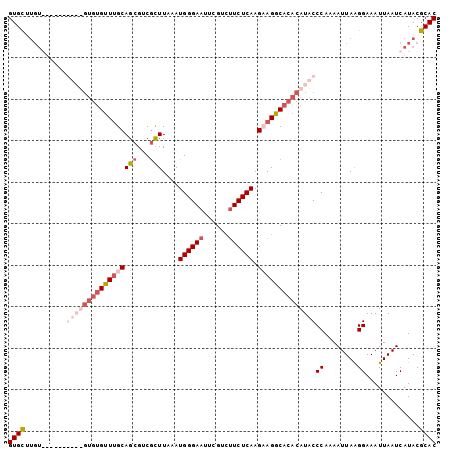
| Location | 5,042,709 – 5,042,812 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -16.10 |
| Energy contribution | -19.73 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5042709 103 - 23771897 GUGCUUGUGUGUGUGUGUGUGUGUUUGCAGCGACGCUUAAAUGGGAAUUUGUGAUCUCAAGAAGGCACACAUACCCAAAAUUAGGGAAAUUAAUCAUACGCAC ......((((((((((((((((((((((......)).....(((((........)))))...)))))))))))(((.......)))........))))))))) ( -31.70) >DroSec_CAF1 249198 89 - 1 GUGCUUGU--------------GUUUGCAGCGUCGCUUAAAUGGGAAUUCGUCUUCUCAAGUAGGCACACAUACCCAAAAUUAAGGAAAUUAAUCAUACGCAC ((((.(((--------------(.(((..(.((.(((((..((((((......))))))..))))))).)....((........))....))).)))).)))) ( -20.80) >DroSim_CAF1 270059 97 - 1 GUGCUUGUGUGU------GUGUGUUUGCAGCGUCGUUUAAAUGGGAAUUCGUCUUCUCAAGUAGGCACACAUACCCAAAAUUAAGGAAAUUAAUCAUACGCAC (((((((.((((------(((((((((((((...)))....((((((......)))))).)))))))))))))).))).(((((.....))))).....)))) ( -28.60) >DroEre_CAF1 252638 95 - 1 GUGUG--------UGUGUGUGUGCUGGCAGGGGCAUUUAAAUGGGAAUUCGUCUUCUCAAGAAGGCACACAUACCCAAAAUUAAGGAAAUUAAUCAUACGCAC (((((--------((((((((((((.((....)).......((((((......))))))....)))))))))))((........))...........)))))) ( -29.70) >consensus GUGCUUGU__________GUGUGUUUGCAGCGUCGCUUAAAUGGGAAUUCGUCUUCUCAAGAAGGCACACAUACCCAAAAUUAAGGAAAUUAAUCAUACGCAC ((((..........(((((((((((((((((...)))....((((((......)))))).))))))))))))))((........)).............)))) (-16.10 = -19.73 + 3.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:13 2006