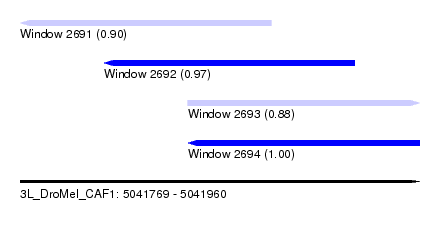
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,041,769 – 5,041,960 |
| Length | 191 |
| Max. P | 0.998600 |

| Location | 5,041,769 – 5,041,889 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.25 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5041769 120 - 23771897 AUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAGCAAAGCUUCUUUAAAGCCAAAAAUAAACUAAUGUGAACGU ..(((((((((((((((((((...(((......)))..((....)).......))))))))))))).....((......))..)))))).....................(((....))) ( -27.70) >DroSec_CAF1 248263 120 - 1 AUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAGCAAAGCUUCUUCAAAGCCAAAAAUAAACUAAUGUGGAUGU ..(((((((((((((((((((...(((......)))..((....)).......))))))))))))).....((......))..)))))).......(((...((......)).))).... ( -28.30) >DroSim_CAF1 269124 120 - 1 AUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAGCAAAGCUUCUUUAAAGCCAAAAAUAAACUAGUGUGGAUGU ..(((((((((((((((((((...(((......)))..((....)).......))))))))))))).....((......))..)))))).......(((...((......)).))).... ( -28.60) >DroEre_CAF1 251702 120 - 1 AUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAGCAAAGCUCCUUAAAAGCCAAAAAUAAACUAAUGUGCAUGU ......((((((............(((......))).(((((.(((.......((((.((....)).))))((......))..))))))))...)))))).................... ( -25.00) >DroYak_CAF1 258066 120 - 1 AUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAUGCAAAGCUCCUUUAAAGCCAAAAAUAAACUAAUGUGCAUGU ......((((((............(((......)))((((((.(((.......((((.((....)).))))(((....)))..)))))))))..)))))).................... ( -26.60) >consensus AUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAGCAAAGCUUCUUUAAAGCCAAAAAUAAACUAAUGUGCAUGU ..(((((((((((((((((((...(((......)))..((....)).......))))))))))))).....((......))..))))))............................... (-25.64 = -26.04 + 0.40)

| Location | 5,041,809 – 5,041,929 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5041809 120 - 23771897 CCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAG ....(((((..((.((((.((((.....))))..)))).))..)))))(((((((((((((...(((......)))..((....)).......))))))))))))).............. ( -32.70) >DroSec_CAF1 248303 120 - 1 CCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAG ....(((((..((.((((.((((.....))))..)))).))..)))))(((((((((((((...(((......)))..((....)).......))))))))))))).............. ( -32.70) >DroSim_CAF1 269164 120 - 1 CCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAG ....(((((..((.((((.((((.....))))..)))).))..)))))(((((((((((((...(((......)))..((....)).......))))))))))))).............. ( -32.70) >DroEre_CAF1 251742 120 - 1 CCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAG ....(((((..((.((((.((((.....))))..)))).))..)))))(((((((((((((...(((......)))..((....)).......))))))))))))).............. ( -32.70) >DroYak_CAF1 258106 120 - 1 CCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAUG ....(((((..((.((((.((((.....))))..)))).))..)))))(((((((((((((...(((......)))..((....)).......))))))))))))).............. ( -32.70) >consensus CCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAGGGCGCUAAAACAAUUUACGGUUAGCCAUAAAGCAAUUAAG ....(((((..((.((((.((((.....))))..)))).))..)))))(((((((((((((...(((......)))..((....)).......))))))))))))).............. (-32.70 = -32.70 + -0.00)

| Location | 5,041,849 – 5,041,960 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.86 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.26 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5041849 111 + 23771897 CUUUGCCCAUAUCGGCGGUUCCACAAUUAGCCAACUUCAUUAUGCCUGCUGCUAAUUGCAAUAACAAAACAAAGUCUAGGCAAAUGUGUAAGCCAUAAAUGAUAACCGGCA--------- ...((((..(((((((((((........)))).((..((((.((((((.(((.....)))...((........)).)))))))))).))..)))......))))...))))--------- ( -23.10) >DroSec_CAF1 248343 111 + 1 CUUUGCCCAUAUCGGCGGUUCCACAAUUAGCCAACUUCAUUAUGCCUGCUGCUAAUUGCAAUAACAAAACAAAGUCUAGGCAAAUGUGUAAGCCAUAAAUGAUAACCGGCA--------- ...((((..(((((((((((........)))).((..((((.((((((.(((.....)))...((........)).)))))))))).))..)))......))))...))))--------- ( -23.10) >DroSim_CAF1 269204 111 + 1 CUUUGCCCAUAUCGGCGGUUCCACAAUUAGCCAACUUCAUUAUGCCUGCUGCUAAUUGCAAUAACAAAACAAAGUCUAGGCAAAUGUGUAAGCCAUAAAUGAUAACCGGCA--------- ...((((..(((((((((((........)))).((..((((.((((((.(((.....)))...((........)).)))))))))).))..)))......))))...))))--------- ( -23.10) >DroEre_CAF1 251782 111 + 1 CUUUGCCCAUAUCGGCGGUUCCACAAUUAGCCAACUUCAUUAUGCCUGCUGCUAAUUGCAAUAACAAAACAAAGUCUAGGCAAAUGUGUAAGCCAUAAAUGAUAACCGGCA--------- ...((((..(((((((((((........)))).((..((((.((((((.(((.....)))...((........)).)))))))))).))..)))......))))...))))--------- ( -23.10) >DroYak_CAF1 258146 120 + 1 CUUUGCCCAUAUCGGCGGUUCCACAAUUAGCCAACUUCAUUAUGCCUGCUGCUAAUUGCAAUAACAAAACAAAGUCUAGGCAAAUGUGUAAGCCAUAAAUGAUAACCGGCAACUGGCAAC ..(((((......(((((((........)))).((..((((.((((((.(((.....)))...((........)).)))))))))).))..))).............(....).))))). ( -26.10) >consensus CUUUGCCCAUAUCGGCGGUUCCACAAUUAGCCAACUUCAUUAUGCCUGCUGCUAAUUGCAAUAACAAAACAAAGUCUAGGCAAAUGUGUAAGCCAUAAAUGAUAACCGGCA_________ ...((((..(((((((((((........)))).((..((((.((((((.(((.....)))...((........)).)))))))))).))..)))......))))...))))......... (-23.26 = -23.26 + -0.00)


| Location | 5,041,849 – 5,041,960 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.86 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -32.34 |
| Energy contribution | -32.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5041849 111 - 23771897 ---------UGCCGGUUAUCAUUUAUGGCUUACACAUUUGCCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAG ---------((((.((.(((......(((...((((.(((((..(((((..((.((((.((((.....))))..)))).))..)))))))).)).)))).....)))))))).))))... ( -32.30) >DroSec_CAF1 248343 111 - 1 ---------UGCCGGUUAUCAUUUAUGGCUUACACAUUUGCCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAG ---------((((.((.(((......(((...((((.(((((..(((((..((.((((.((((.....))))..)))).))..)))))))).)).)))).....)))))))).))))... ( -32.30) >DroSim_CAF1 269204 111 - 1 ---------UGCCGGUUAUCAUUUAUGGCUUACACAUUUGCCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAG ---------((((.((.(((......(((...((((.(((((..(((((..((.((((.((((.....))))..)))).))..)))))))).)).)))).....)))))))).))))... ( -32.30) >DroEre_CAF1 251782 111 - 1 ---------UGCCGGUUAUCAUUUAUGGCUUACACAUUUGCCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAG ---------((((.((.(((......(((...((((.(((((..(((((..((.((((.((((.....))))..)))).))..)))))))).)).)))).....)))))))).))))... ( -32.30) >DroYak_CAF1 258146 120 - 1 GUUGCCAGUUGCCGGUUAUCAUUUAUGGCUUACACAUUUGCCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAG .(((((.((((.(((((.((((...(((((.((.(((((((((.(((........))).((((.....)))).))))).))))..)).)))))...))))))))).))))...))))).. ( -37.70) >consensus _________UGCCGGUUAUCAUUUAUGGCUUACACAUUUGCCUAGACUUUGUUUUGUUAUUGCAAUUAGCAGCAGGCAUAAUGAAGUUGGCUAAUUGUGGAACCGCCGAUAUGGGCAAAG .........((((.((.(((......(((...((((.(((((..(((((..((.((((.((((.....))))..)))).))..)))))))).)).)))).....)))))))).))))... (-32.34 = -32.34 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:09 2006