| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,041,423 – 5,041,581 |
| Length | 158 |
| Max. P | 0.926049 |

| Location | 5,041,423 – 5,041,542 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.24 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5041423 119 + 23771897 GUCA-AAUCGAAUUUCGCAUUUGCUGGGCCAAAAACAUUAUCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUG (.((-(((((.....)).))))))...(((((...((((((.......))))))......((((((((.....))))))))..)))))....(((((((((..........))))))))) ( -24.60) >DroSec_CAF1 247891 119 + 1 GUCA-AAUCGAAUUUCGCAUUUGCUGGGCCAAAAACAUUAUCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUG (.((-(((((.....)).))))))...(((((...((((((.......))))))......((((((((.....))))))))..)))))....(((((((((..........))))))))) ( -24.60) >DroSim_CAF1 268753 119 + 1 GUCA-AAUCGAAUUUCGCAUUUGCUGGGCCAAAAACAUUAUCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUG (.((-(((((.....)).))))))...(((((...((((((.......))))))......((((((((.....))))))))..)))))....(((((((((..........))))))))) ( -24.60) >DroEre_CAF1 251339 119 + 1 GUCA-AAUCGAAUUUCGCAUUUGCUGGGCCAAAAACAUUAUCGCAUAAAUGAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUG (.((-(((((.....)).))))))...(((((...((((((.......))))))......((((((((.....))))))))..)))))....(((((((((..........))))))))) ( -24.70) >DroYak_CAF1 257728 120 + 1 UUAAAAAUCGAAUUUCGCAUUUGCUGGGCCAAAAACAUUAUCGCAUAAAUGAUGACAUCUACUGGCAUUUAAAAUGUCUAUCAUUGGUGGAACAUUUCAUUAAAAUAAACCAAUGGAAUG ...........(((((((....))(((((((......((((((......)))))).......)))).....((((((((((.....)))).))))))............)))..))))). ( -20.52) >consensus GUCA_AAUCGAAUUUCGCAUUUGCUGGGCCAAAAACAUUAUCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUG ............((((((....))...(((((...((((((.......))))))......((((((((.....))))))))..)))))))))(((((((((..........))))))))) (-22.40 = -22.24 + -0.16)


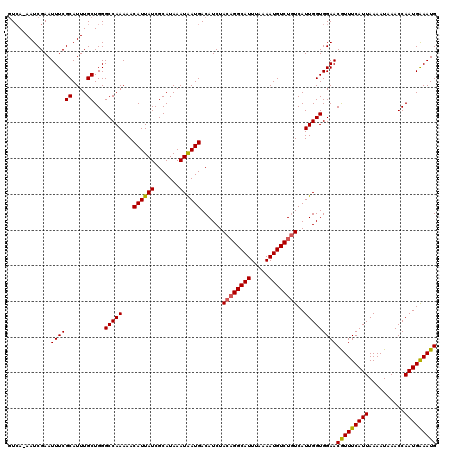
| Location | 5,041,462 – 5,041,581 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5041462 119 + 23771897 UCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUGGCUUACUUUGGUUCCCUUUUCGCUUACACGGAA-AAAAUA ..((...........((((.((((((((.....))))))))....))))...(((((((((..........)))))))))))..............((((((......)))))-)..... ( -23.60) >DroSec_CAF1 247930 120 + 1 UCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUGGCUUACUUUGGUUCCCUUUUCGCUUACACGGAACAAAAUA ..((...........((((.((((((((.....))))))))....))))...(((((((((..........)))))))))))........(((((..............)))))...... ( -25.04) >DroSim_CAF1 268792 120 + 1 UCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUGGCUUACUUUAGUUCCCUUUUCGCUUACACGGAAGAAAAUA ..((...........((((.((((((((.....))))))))....))))...(((((((((..........)))))))))))..............((((((......))))))...... ( -23.30) >DroEre_CAF1 251378 120 + 1 UCGCAUAAAUGAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUGGCUUACUUUGGUUGCCUUUCGGCUUACACAAGAAAAAAUA ..((...........((((.((((((((.....))))))))....))))...(((((((((..........)))))))))))....((((((.(((....)))..)).))))........ ( -26.70) >DroYak_CAF1 257768 120 + 1 UCGCAUAAAUGAUGACAUCUACUGGCAUUUAAAAUGUCUAUCAUUGGUGGAACAUUUCAUUAAAAUAAACCAAUGGAAUGGAUUACUUUGGUUCUCUUUUCGCUUACACACGAAAAAAUA ((((...(((((((((((...............))))).)))))).))))..(((((((((..........)))))))))((..((....))..))((((((........)))))).... ( -21.86) >consensus UCGCAUAAAUAAUGACAUCUACAGGCAUUUAAAAUGUCUGUCAUUGGUGGAACGUUUCAUUAAAAUAAACCAAUGAAAUGGCUUACUUUGGUUCCCUUUUCGCUUACACGGAAAAAAAUA ..((......(((((((......(((((.....))))))))))))(((((..(((((((((..........)))))))))..)))))..............))................. (-20.56 = -20.20 + -0.36)

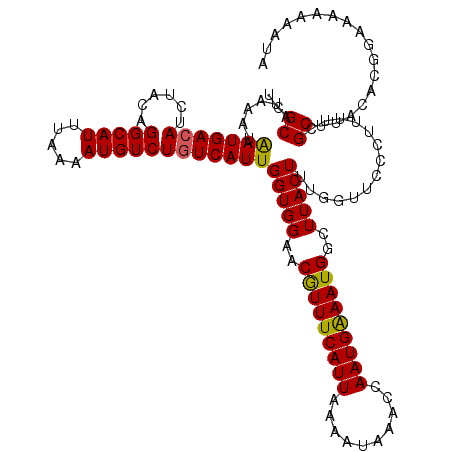
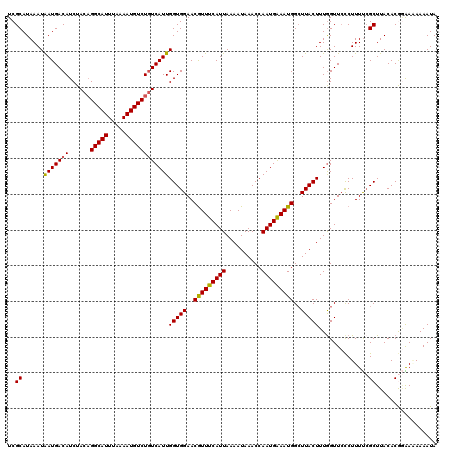
| Location | 5,041,462 – 5,041,581 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.66 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.926049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5041462 119 - 23771897 UAUUUU-UUCCGUGUAAGCGAAAAGGGAACCAAAGUAAGCCAUUUCAUUGGUUUAUUUUAAUGAAACGUUCCACCAAUGACAGACAUUUUAAAUGCCUGUAGAUGUCAUUAUUUAUGCGA ......-...((((((((.......(((((.(((((((((((......)))))))))))........)))))...(((((((.(((...........)))...))))))).)))))))). ( -27.90) >DroSec_CAF1 247930 120 - 1 UAUUUUGUUCCGUGUAAGCGAAAAGGGAACCAAAGUAAGCCAUUUCAUUGGUUUAUUUUAAUGAAACGUUCCACCAAUGACAGACAUUUUAAAUGCCUGUAGAUGUCAUUAUUUAUGCGA ..........((((((((.......(((((.(((((((((((......)))))))))))........)))))...(((((((.(((...........)))...))))))).)))))))). ( -27.90) >DroSim_CAF1 268792 120 - 1 UAUUUUCUUCCGUGUAAGCGAAAAGGGAACUAAAGUAAGCCAUUUCAUUGGUUUAUUUUAAUGAAACGUUCCACCAAUGACAGACAUUUUAAAUGCCUGUAGAUGUCAUUAUUUAUGCGA ..........((((((((.......(((((((((((((((((......)))))))))))).......)))))...(((((((.(((...........)))...))))))).)))))))). ( -30.21) >DroEre_CAF1 251378 120 - 1 UAUUUUUUCUUGUGUAAGCCGAAAGGCAACCAAAGUAAGCCAUUUCAUUGGUUUAUUUUAAUGAAACGUUCCACCAAUGACAGACAUUUUAAAUGCCUGUAGAUGUCAUCAUUUAUGCGA .........(((((((((((....)))....(((((((((((......))))))))))).((((...(((........))).(((((((...........))))))).)))))))))))) ( -29.70) >DroYak_CAF1 257768 120 - 1 UAUUUUUUCGUGUGUAAGCGAAAAGAGAACCAAAGUAAUCCAUUCCAUUGGUUUAUUUUAAUGAAAUGUUCCACCAAUGAUAGACAUUUUAAAUGCCAGUAGAUGUCAUCAUUUAUGCGA ..(((((((((......))))))))).....(((((((.(((......))).)))))))..........((((..((((((.(((((((...........)))))))))))))..)).)) ( -25.50) >consensus UAUUUUUUUCCGUGUAAGCGAAAAGGGAACCAAAGUAAGCCAUUUCAUUGGUUUAUUUUAAUGAAACGUUCCACCAAUGACAGACAUUUUAAAUGCCUGUAGAUGUCAUUAUUUAUGCGA ..........((((((((.((....(((((.(((((((((((......)))))))))))........)))))..........(((((((...........))))))).)).)))))))). (-22.38 = -22.66 + 0.28)

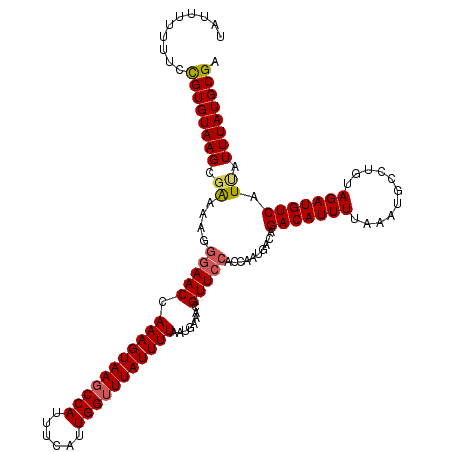
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:05 2006