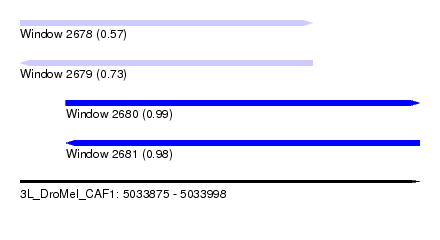
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,033,875 – 5,033,998 |
| Length | 123 |
| Max. P | 0.989035 |

| Location | 5,033,875 – 5,033,965 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -11.86 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5033875 90 + 23771897 CCGAGAUACUCAAC------AAAAAAAAUUGAAUAAAAAUGCACU-CGGCGUGCAGGAGCAAAGGGCAAGAUGUUGUGUA--GCAGCUAGUAGCUGCUA ......(((.((((------(..................(((((.-....)))))...((.....))....))))).)))--(((((.....))))).. ( -23.50) >DroPse_CAF1 305411 74 + 1 GGGGGAUGCCCG--------AAA--AAAUUGAAUAAAAAUGCACUGCAGCGUGCAGCGAGAGAGAG--AGAGGGAG------------AGAAUCU-CUC (((((((.(((.--------...--..............(((((......))))).(....)....--...)))..------------...))))-))) ( -18.40) >DroSec_CAF1 240463 88 + 1 CCGAGAUCCUCAAC------AAA--AAAUUGAAUAAAAAUGCACU-CGGCGUGCAGGAGCAAAGGGCAAGAUGUUGUGUA--GCAGCUAGUAGCUGCUA ((....(((((((.------...--...)))).......(((((.-....)))))))).....))((((....)))).((--(((((.....))))))) ( -22.80) >DroSim_CAF1 261321 88 + 1 CCGAGAUCCUCAAC------AAA--AAAUUGAAUAAAAAUGCACU-CGGCGUGCAGGAGCAAAGGGCAAGAUGUUGUGUA--GCAGCUAGUAGCUGCUA ((....(((((((.------...--...)))).......(((((.-....)))))))).....))((((....)))).((--(((((.....))))))) ( -22.80) >DroEre_CAF1 243780 85 + 1 CCGAGAUCCACAAC------AAA--AAAUUGAAUAAAAAUGCACU-CGGCGUGCAGGAGCAAAAGGCGAGAUGUUGUA-----CAGCUAGUAGCUGCUA .........(((((------(..--..(((.......)))...((-((.(.(((....)))...).)))).)))))).-----((((.....))))... ( -19.40) >DroYak_CAF1 249909 96 + 1 CCGAGAUCCUCAACCAAAAAAAA--AAAUUGAAUAAAAAUGCACU-CGCCGUGCAGGAGCAAAAGGCGAGAUGUUGUGUAGAGCAGCCAGUAGCUGCUA ..(((...)))............--...............(((((-((((.(((....)))...)))))).))).......((((((.....)))))). ( -26.50) >consensus CCGAGAUCCUCAAC______AAA__AAAUUGAAUAAAAAUGCACU_CGGCGUGCAGGAGCAAAGGGCAAGAUGUUGUGUA__GCAGCUAGUAGCUGCUA (((((...)))............................(((((......))))))).........................(((((.....))))).. (-11.86 = -12.57 + 0.71)

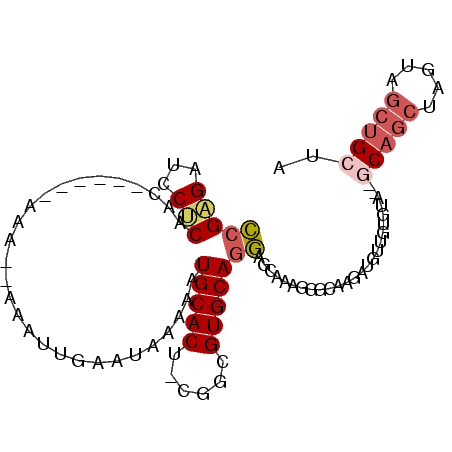
| Location | 5,033,875 – 5,033,965 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -11.14 |
| Energy contribution | -12.85 |
| Covariance contribution | 1.71 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5033875 90 - 23771897 UAGCAGCUACUAGCUGC--UACACAACAUCUUGCCCUUUGCUCCUGCACGCCG-AGUGCAUUUUUAUUCAAUUUUUUUU------GUUGAGUAUCUCGG (((((((.....)))))--)).................(((....)))..(((-((........((((((((.......------)))))))).))))) ( -23.30) >DroPse_CAF1 305411 74 - 1 GAG-AGAUUCU------------CUCCCUCU--CUCUCUCUCGCUGCACGCUGCAGUGCAUUUUUAUUCAAUUU--UUU--------CGGGCAUCCCCC (((-(((....------------.....)))--))).....((((((.....))))))................--...--------.(((....))). ( -17.50) >DroSec_CAF1 240463 88 - 1 UAGCAGCUACUAGCUGC--UACACAACAUCUUGCCCUUUGCUCCUGCACGCCG-AGUGCAUUUUUAUUCAAUUU--UUU------GUUGAGGAUCUCGG (((((((.....)))))--)).............((...(.((((((((....-.))))).......(((((..--...------)))))))).)..)) ( -23.00) >DroSim_CAF1 261321 88 - 1 UAGCAGCUACUAGCUGC--UACACAACAUCUUGCCCUUUGCUCCUGCACGCCG-AGUGCAUUUUUAUUCAAUUU--UUU------GUUGAGGAUCUCGG (((((((.....)))))--)).............((...(.((((((((....-.))))).......(((((..--...------)))))))).)..)) ( -23.00) >DroEre_CAF1 243780 85 - 1 UAGCAGCUACUAGCUG-----UACAACAUCUCGCCUUUUGCUCCUGCACGCCG-AGUGCAUUUUUAUUCAAUUU--UUU------GUUGUGGAUCUCGG ..(((((.....))))-----)((((((....((.....))...(((((....-.)))))..............--..)------)))))......... ( -19.00) >DroYak_CAF1 249909 96 - 1 UAGCAGCUACUGGCUGCUCUACACAACAUCUCGCCUUUUGCUCCUGCACGGCG-AGUGCAUUUUUAUUCAAUUU--UUUUUUUUGGUUGAGGAUCUCGG .((((((.....))))))........((.((((((...(((....))).))))-)))).(((((((..(((...--......)))..)))))))..... ( -25.90) >consensus UAGCAGCUACUAGCUGC__UACACAACAUCUUGCCCUUUGCUCCUGCACGCCG_AGUGCAUUUUUAUUCAAUUU__UUU______GUUGAGGAUCUCGG ..(((((.....)))))......((((.....((.....))...(((((......))))).........................)))).......... (-11.14 = -12.85 + 1.71)

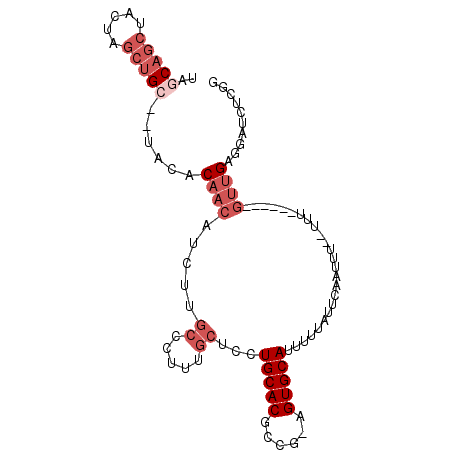

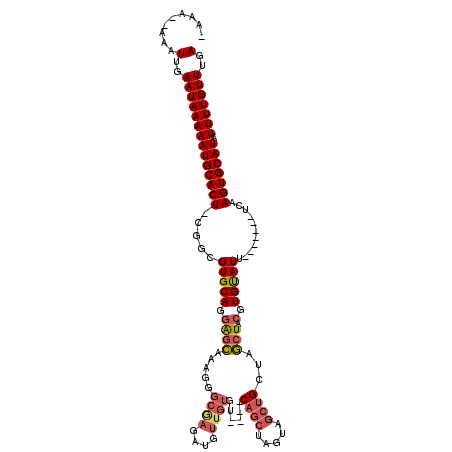
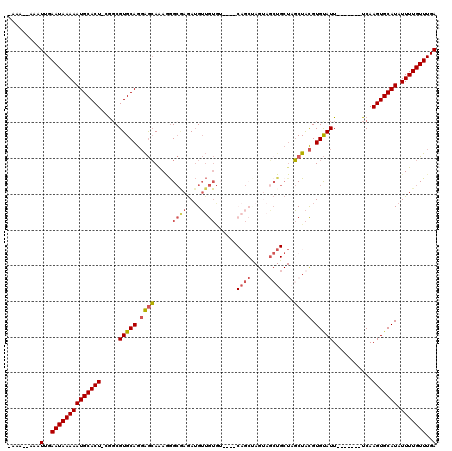
| Location | 5,033,889 – 5,033,998 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.99 |
| Covariance contribution | 0.99 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.85 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5033889 109 + 23771897 -AAAAAAAAUUGAAUAAAAAUGCACU-CGGCGUGCAGGAGCAAAGGGCAAGAUGUUGUGUA--GCAGCUAGUAGCUGCUAGCUACGUGUAUU-------UCAAGUGCAUAUUUUGUUUGA -........(..((((((((((((((-.((.(((((.((((.....((((....)))).((--(((((.....)))))))))).).))))).-------)).))))))).)))))))..) ( -38.60) >DroPse_CAF1 305423 95 + 1 -AAA--AAAUUGAAUAAAAAUGCACUGCAGCGUGCAGCGAGAGAGAG--AGAGGGAG------------AGAAUCU-CUCUCUGUGUGCAUU-------UCAAGUGCAUAUUUUGUUUGA -...--...(..((((((((((((((.....(((((.((..((((((--(((.....------------....)))-)))))).))))))).-------...))))))).)))))))..) ( -34.80) >DroSim_CAF1 261335 107 + 1 -AAA--AAAUUGAAUAAAAAUGCACU-CGGCGUGCAGGAGCAAAGGGCAAGAUGUUGUGUA--GCAGCUAGUAGCUGCUAGCUACGUGUAUU-------UCAAGUGCAUAUUUUGUUUGA -...--...(..((((((((((((((-.((.(((((.((((.....((((....)))).((--(((((.....)))))))))).).))))).-------)).))))))).)))))))..) ( -38.60) >DroEre_CAF1 243794 111 + 1 -AAA--AAAUUGAAUAAAAAUGCACU-CGGCGUGCAGGAGCAAAAGGCGAGAUGUUGUA-----CAGCUAGUAGCUGCUAGCUACGUGUAUUUCGAAUUUCAAGUGCAUAUUUUGUUUGA -...--...(..((((((((((((((-.(.(.(((....)))...).)(((((...(((-----((((((((....))))))....))))).....))))).))))))).)))))))..) ( -32.10) >DroYak_CAF1 249928 110 + 1 AAAA--AAAUUGAAUAAAAAUGCACU-CGCCGUGCAGGAGCAAAAGGCGAGAUGUUGUGUAGAGCAGCCAGUAGCUGCUAGCUACGUGUAUU-------UCAAGUGCAUAUUUUGUUUGA ....--...(..((((((((((((((-.(((.(((....)))...)))((((((...(((((((((((.....))))))..)))))..))))-------)).))))))).)))))))..) ( -39.90) >DroAna_CAF1 251063 89 + 1 -AAA--AAAUUGAAUAAAAAUGCACU-UGGCGUGCAGGAGCAAA-GGCGAAAUGUUGUGU-------------------AGCUACGUGUAUU-------UCAAGUGCAUAUUUUGUUUGA -...--...(..((((((((((((((-(((.(((((.((((..(-.((((....)))).)-------------------.))).).))))).-------)))))))))).)))))))..) ( -30.80) >consensus _AAA__AAAUUGAAUAAAAAUGCACU_CGGCGUGCAGGAGCAAAGGGCGAGAUGUUGUGU____CAGCUAGUAGCUGCUAGCUACGUGUAUU_______UCAAGUGCAUAUUUUGUUUGA .........(..((((((((((((((.....(((((.((((.....((((....))))......((((.....))))...))).).)))))...........))))))).)))))))..) (-19.00 = -19.99 + 0.99)

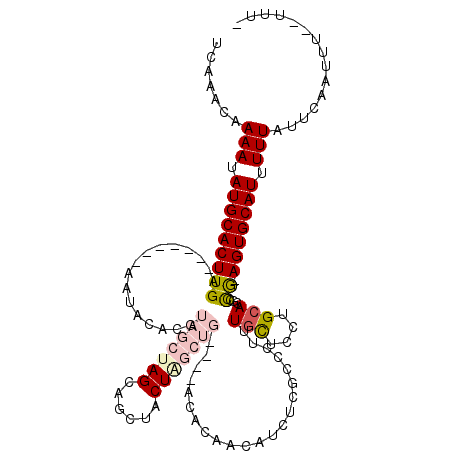
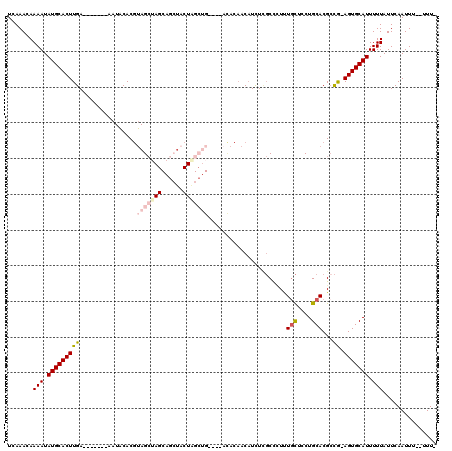
| Location | 5,033,889 – 5,033,998 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -12.02 |
| Energy contribution | -13.43 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5033889 109 - 23771897 UCAAACAAAAUAUGCACUUGA-------AAUACACGUAGCUAGCAGCUACUAGCUGC--UACACAACAUCUUGCCCUUUGCUCCUGCACGCCG-AGUGCAUUUUUAUUCAAUUUUUUUU- .......(((.(((((((((.-------.......((((((((......))))))))--...................(((....)))...))-))))))).)))..............- ( -27.10) >DroPse_CAF1 305423 95 - 1 UCAAACAAAAUAUGCACUUGA-------AAUGCACACAGAGAG-AGAUUCU------------CUCCCUCU--CUCUCUCUCGCUGCACGCUGCAGUGCAUUUUUAUUCAAUUU--UUU- ......(((((.((.....((-------(((((....((((((-(((....------------.....)))--))))))...(((((.....))))))))))))....))))))--)..- ( -25.40) >DroSim_CAF1 261335 107 - 1 UCAAACAAAAUAUGCACUUGA-------AAUACACGUAGCUAGCAGCUACUAGCUGC--UACACAACAUCUUGCCCUUUGCUCCUGCACGCCG-AGUGCAUUUUUAUUCAAUUU--UUU- .......(((.(((((((((.-------.......((((((((......))))))))--...................(((....)))...))-))))))).))).........--...- ( -27.10) >DroEre_CAF1 243794 111 - 1 UCAAACAAAAUAUGCACUUGAAAUUCGAAAUACACGUAGCUAGCAGCUACUAGCUG-----UACAACAUCUCGCCUUUUGCUCCUGCACGCCG-AGUGCAUUUUUAUUCAAUUU--UUU- .......(((.(((((((((................(((((((......)))))))-----...........((....(((....))).))))-))))))).))).........--...- ( -23.70) >DroYak_CAF1 249928 110 - 1 UCAAACAAAAUAUGCACUUGA-------AAUACACGUAGCUAGCAGCUACUGGCUGCUCUACACAACAUCUCGCCUUUUGCUCCUGCACGGCG-AGUGCAUUUUUAUUCAAUUU--UUUU .......(((.((((((....-------.......((((..((((((.....))))))))))........(((((...(((....))).))))-))))))).))).........--.... ( -32.50) >DroAna_CAF1 251063 89 - 1 UCAAACAAAAUAUGCACUUGA-------AAUACACGUAGCU-------------------ACACAACAUUUCGCC-UUUGCUCCUGCACGCCA-AGUGCAUUUUUAUUCAAUUU--UUU- .......(((.(((((((((.-------.......((....-------------------))..........((.-..(((....))).))))-))))))).))).........--...- ( -15.70) >consensus UCAAACAAAAUAUGCACUUGA_______AAUACACGUAGCUAGCAGCUACUAGCUG____ACACAACAUCUCGCCCUUUGCUCCUGCACGCCG_AGUGCAUUUUUAUUCAAUUU__UUU_ .......(((.(((((((((................(((((((......)))))))......................(((....)))...)).))))))).)))............... (-12.02 = -13.43 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:56 2006