
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,013,590 – 5,013,699 |
| Length | 109 |
| Max. P | 0.582704 |

| Location | 5,013,590 – 5,013,699 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -22.73 |
| Energy contribution | -21.98 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
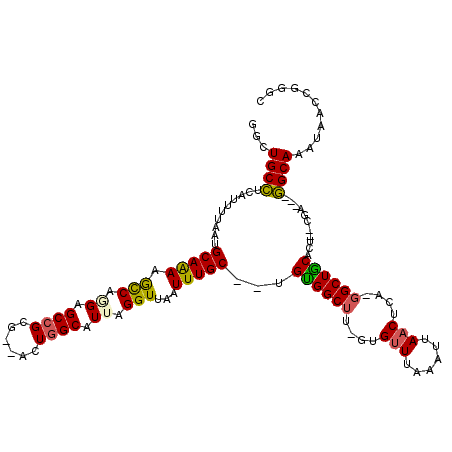
>3L_DroMel_CAF1 5013590 109 - 23771897 GGCUGCCUUAUUUUAAUGCAAACGCCAGGAGCCGCG--ACUGGCAUUAGGUUAAUUUGC--UGCGGCUU-GUGUUUAAAUUAACUCA-GGCUGCACU--CGA---GGCAAAUAACCGGGC ....((((...............(((((........--.)))))....(((((.(((((--((((((((-(.(((......))).))-))))))...--...---))))))))))))))) ( -38.30) >DroPse_CAF1 294420 112 - 1 UGCUGCUUCCAUUUAAUGCAGAAACCAUGAGCCGCGGAGCUGGCAUUAGGUUAAUUUGCCGUGUGGCUUCGUGUUUAAAUUAACUCUCGGCUACACUCUCGA---AGCAAAUAUC----- ...((((((........(((((((((.((((((((...)).))).)))))))..))))).((((((((..(.(((......))).)..))))))))....))---))))......----- ( -33.80) >DroEre_CAF1 237448 109 - 1 GGCUGCCUUAUUUUAAUGCAAACGCCAGGAGCCGCG--ACUAGCAUUAGGUUAAUUUGC--UGUGGCUU-GUGCUUAAAUUAACUCA-GGCUGCACU--CGA---GGCAAAUAACCGGGC ....((((.....((((((...(((........)))--....))))))(((((.(((((--((..((((-(.(..........).))-)))..)...--...---))))))))))))))) ( -30.70) >DroYak_CAF1 242942 109 - 1 UGCUGCCUUAUUUUAAUGCAAAUGUCAGGAGCCGCG--ACUGGCAUUAGGUUAAUUUGC--UGCGGCUU-GUGUUUAAAUUAACUCA-GGCUGCACU--CGA---GGCAAAUAACCGGGC ....((((.....((((((....(((.(....)..)--))..))))))(((((.(((((--((((((((-(.(((......))).))-))))))...--...---))))))))))))))) ( -38.80) >DroAna_CAF1 244453 110 - 1 GCCUGCCUCAUUUUAAUGCAGAGGCCAAGAGCCGCG--ACUGGCAUUAGGUUAAUUUGC--AGUGGCUU-GUGUUUAAAUUAACUCU-CGCUGCCCU--CGAGAAGGCAACU--CCGAGU ((((..(((.......((((((.(((.((.((((..--..)))).)).)))...)))))--)(..((..-(.(((......))).).-.))..)...--.))).))))....--...... ( -32.10) >DroPer_CAF1 266012 112 - 1 UGCUGCUUCCAUUUAAUGCAGAAACCAUGAGCCGCGGAGCUGGCAUUAGGUUAAUUUGCCGUGUGGCUUCGUGUUUAAAUUAACUCUCGGCUACACUCUCGA---AGCAAAUAUC----- ...((((((........(((((((((.((((((((...)).))).)))))))..))))).((((((((..(.(((......))).)..))))))))....))---))))......----- ( -33.80) >consensus GGCUGCCUCAUUUUAAUGCAAAAGCCAGGAGCCGCG__ACUGGCAUUAGGUUAAUUUGC__UGUGGCUU_GUGUUUAAAUUAACUCA_GGCUGCACU__CGA___GGCAAAUAACCGGGC ...((((..........(((((.(((.((.((((......)))).)).)))...)))))...((((((....(((......)))....))))))...........))))........... (-22.73 = -21.98 + -0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:49 2006