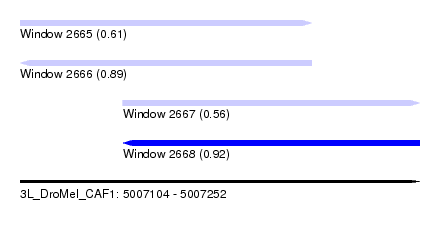
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,007,104 – 5,007,252 |
| Length | 148 |
| Max. P | 0.917863 |

| Location | 5,007,104 – 5,007,212 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -23.44 |
| Energy contribution | -26.05 |
| Covariance contribution | 2.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
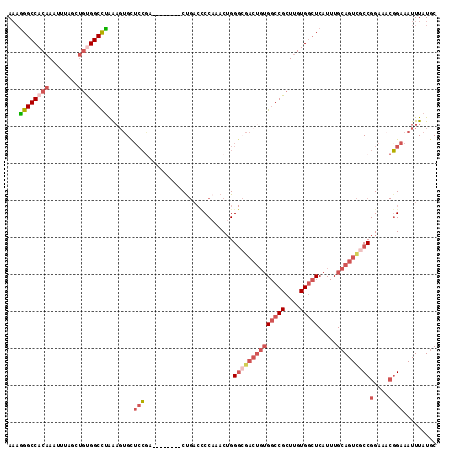
>3L_DroMel_CAF1 5007104 108 + 23771897 GCAUAAAUUUCCGUUUCCGGCGACUGCAAAUGAGCCACAAGCGGCCACAGUCGCCCAGUUUGGGGUCAG--------UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUU ............((((((((((((((....((.(((......))))))))))((((......))))..)--------)))))).))....((((((((......)))))))).... ( -40.10) >DroSec_CAF1 227589 108 + 1 GCAUAAAUUUCCGUUUCCGGCGACUGCAAAUGAGCCACAAGCGGCCACAGUCGCCCAGUUUGGAGUCAG--------UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUU ........(((((.((((((((((((....((.(((......)))))))))))))......))))....--------.))))).......((((((((......)))))))).... ( -38.50) >DroSim_CAF1 247621 86 + 1 G-AUAAAUUUCG--UUCCGGCAACUGCA-AUGAU-CAC-AGCGG-CACAGUC--CCAGUUGGGU---AG--------C-CGAGCA--UUAAGCCA--ACUA---UUGUGGCCU--U .-.........(--(((.(((..((((.-.....-...-.))))-......(--((....))).---.)--------)-))))).--....((((--.(..---..)))))..--. ( -21.00) >DroEre_CAF1 230797 108 + 1 GCAUAAAUUUCCGUUUCCGGCGACUGCAAAUGAGCCACAAGCGGCCACAGUCGCCCAGUUUGGGGUCAG--------UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUU ............((((((((((((((....((.(((......))))))))))((((......))))..)--------)))))).))....((((((((......)))))))).... ( -40.10) >DroYak_CAF1 236205 108 + 1 GCAUAAAUUUCCGUUUCCGGCGACUGCAAAUGAGCCACAAGCGGCCACAGUCGCCCAGUUUGGGGUCAG--------UCGGAGCACUUUAGGCCGCAGCUAAAUUUGUGGCCCUUU ............((((((((((((((....((.(((......))))))))))((((......))))..)--------)))))).))....((((((((......)))))))).... ( -39.70) >DroAna_CAF1 238169 109 + 1 GCAUAAAUUUCCGUUUCCGGCGACUGCAAAUGAGCCACAAGCGGCC-------CCCUGUUUGGGGUCGGCUAGCUAGUAGCUACUCUCCGGGCCGCAACUAAAUUUGUGGCCCUUU (((....((.(((....))).)).))).....(((.((.(((((((-------((((....))))..)))).))).)).))).......(((((((((......)))))))))... ( -39.30) >consensus GCAUAAAUUUCCGUUUCCGGCGACUGCAAAUGAGCCACAAGCGGCCACAGUCGCCCAGUUUGGGGUCAG________UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUU ........(((((.....((((((((....((.(((......))))))))))))).....))))).........................((((((((......)))))))).... (-23.44 = -26.05 + 2.61)

| Location | 5,007,104 – 5,007,212 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -28.87 |
| Energy contribution | -31.07 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
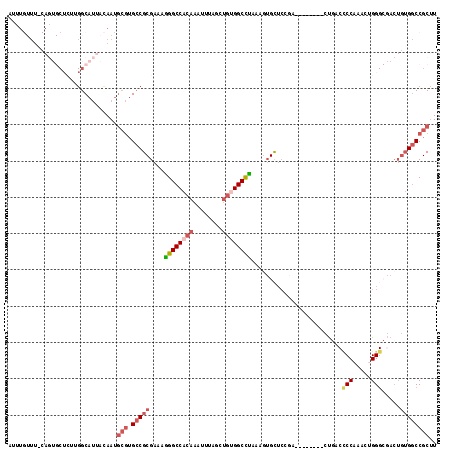
>3L_DroMel_CAF1 5007104 108 - 23771897 AAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA--------CUGACCCCAAACUGGGCGACUGUGGCCGCUUGUGGCUCAUUUGCAGUCGCCGGAAACGGAAAUUUAUGC ...((((((((........)))))))).((((..(((..--------..............((((((((((((((....))))).....)))))))))(....)))).)))).... ( -43.30) >DroSec_CAF1 227589 108 - 1 AAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA--------CUGACUCCAAACUGGGCGACUGUGGCCGCUUGUGGCUCAUUUGCAGUCGCCGGAAACGGAAAUUUAUGC ...((((((((........)))))))).((((..(((..--------..............((((((((((((((....))))).....)))))))))(....)))).)))).... ( -43.30) >DroSim_CAF1 247621 86 - 1 A--AGGCCACAA---UAGU--UGGCUUAA--UGCUCG-G--------CU---ACCCAACUGG--GACUGUG-CCGCU-GUG-AUCAU-UGCAGUUGCCGGAA--CGAAAUUUAU-C .--((((((...---....--))))))..--...(((-.--------..---.(((....))--).(((.(-(.(((-(..-(...)-..)))).)))))..--))).......-. ( -24.40) >DroEre_CAF1 230797 108 - 1 AAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA--------CUGACCCCAAACUGGGCGACUGUGGCCGCUUGUGGCUCAUUUGCAGUCGCCGGAAACGGAAAUUUAUGC ...((((((((........)))))))).((((..(((..--------..............((((((((((((((....))))).....)))))))))(....)))).)))).... ( -43.30) >DroYak_CAF1 236205 108 - 1 AAAGGGCCACAAAUUUAGCUGCGGCCUAAAGUGCUCCGA--------CUGACCCCAAACUGGGCGACUGUGGCCGCUUGUGGCUCAUUUGCAGUCGCCGGAAACGGAAAUUUAUGC ...(((((((((........((((((...(((((((.(.--------.((....))..).)))).)))..)))))))))))))))....(((....(((....))).......))) ( -42.50) >DroAna_CAF1 238169 109 - 1 AAAGGGCCACAAAUUUAGUUGCGGCCCGGAGAGUAGCUACUAGCUAGCCGACCCCAAACAGGG-------GGCCGCUUGUGGCUCAUUUGCAGUCGCCGGAAACGGAAAUUUAUGC ...(((((((((........(((((.(((....((((.....)))).))).((((.....)))-------)))))))))))))))....(((....(((....))).......))) ( -46.10) >consensus AAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA________CUGACCCCAAACUGGGCGACUGUGGCCGCUUGUGGCUCAUUUGCAGUCGCCGGAAACGGAAAUUUAUGC ...((((((((........)))))))).......(((........................((((((((((((((....))))).....)))))))))(....))))......... (-28.87 = -31.07 + 2.20)

| Location | 5,007,142 – 5,007,252 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.22 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -20.63 |
| Energy contribution | -21.85 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
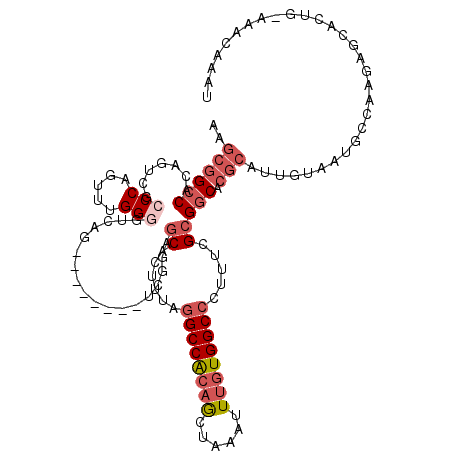
>3L_DroMel_CAF1 5007142 110 + 23771897 AAGCGGCCACAGUCGCCCAGUUUGGGGUCAG--------UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUUCGCGGCACGCAUUGUAAUGCCGAGAGCACUGCAAACAAAU ..(((((....)))))...(((((.((((..--------((((.(((.....((((((((......)))))))).....(((...)))..)))....))))..).))).))))).... ( -37.10) >DroSec_CAF1 227627 110 + 1 AAGCGGCCACAGUCGCCCAGUUUGGAGUCAG--------UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUUCGCGGCACGCAUUGUAAUGCCAAGAGCACUGCAAACAAAU ..((((((.((((.((((...(((....)))--------..)).)))))...((((((((......)))))))).....).))).))).(((((.(((.....))).)))))...... ( -35.50) >DroSim_CAF1 247654 87 + 1 -AGCGG-CACAGUC--CCAGUUGGGU---AG--------C-CGAGCA--UUAAGCCA--ACUA---UUGUGGCCU--UCGCGCCC------UUGAUGCNNNNNNNNNNNNNNNNNNNN -..(((-(.....(--((....))).---.)--------)-)).(((--(((((...--((..---..))(((..--....))))------))))))).................... ( -20.50) >DroEre_CAF1 230835 110 + 1 AAGCGGCCACAGUCGCCCAGUUUGGGGUCAG--------UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUUCGCGGCACGCAUUGUAAUGCCAGGAGCACUGGAAACAUAU ..((((((......((((......))))..(--------(..(((.......((((((((......)))))))))))..))))).)))..(((....((((.....))))..)))... ( -36.61) >DroYak_CAF1 236243 110 + 1 AAGCGGCCACAGUCGCCCAGUUUGGGGUCAG--------UCGGAGCACUUUAGGCCGCAGCUAAAUUUGUGGCCCUUUCGCGGCACGCAUUGUAAUGCCAAGAGCACUGUAAACAAAA ..(((((....)))))...(((((.((((.(--------((((((.......((((((((......))))))))..))).))))..((((....)))).....).))).))))).... ( -32.70) >DroAna_CAF1 238207 97 + 1 AAGCGGCC-------CCCUGUUUGGGGUCGGCUAGCUAGUAGCUACUCUCCGGGCCGCAACUAAAUUUGUGGCCCUUUCGCGGCACGCAUUACAAUGCCGGCCG-------------- ...(((((-------((......)))))))((((((.....))))......(((((((((......)))))))))....))(((..((((....))))..))).-------------- ( -43.20) >consensus AAGCGGCCACAGUCGCCCAGUUUGGGGUCAG________UCGGAGCACUUUAGGCCACAGCUAAAUUUGUGGCCCUUUCGCGGCACGCAUUGUAAUGCCAAGAGCACUG_AAACAAAU ..((((((.......(((.....)))..................((......((((((((......)))))))).....))))).))).............................. (-20.63 = -21.85 + 1.22)


| Location | 5,007,142 – 5,007,252 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.22 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -22.59 |
| Energy contribution | -23.90 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
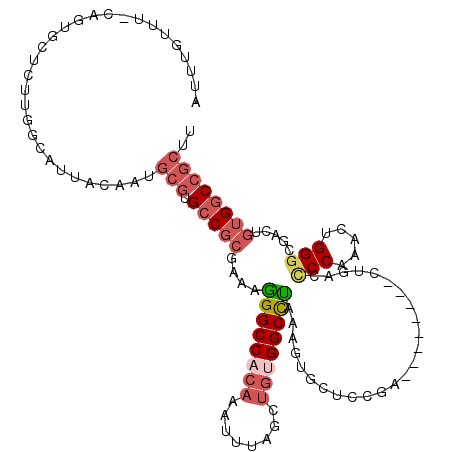
>3L_DroMel_CAF1 5007142 110 - 23771897 AUUUGUUUGCAGUGCUCUCGGCAUUACAAUGCGUGCCGCGAAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA--------CUGACCCCAAACUGGGCGACUGUGGCCGCUU ......(((.(((((.....))))).))).(((.(((((....((((((((........))))))))..(((((((.(.--------.((....))..).)))).))))))))))).. ( -39.70) >DroSec_CAF1 227627 110 - 1 AUUUGUUUGCAGUGCUCUUGGCAUUACAAUGCGUGCCGCGAAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA--------CUGACUCCAAACUGGGCGACUGUGGCCGCUU ......(((.(((((.....))))).))).(((.(((((....((((((((........))))))))..(((((((.(.--------.((....))..).)))).))))))))))).. ( -39.70) >DroSim_CAF1 247654 87 - 1 NNNNNNNNNNNNNNNNNNNNGCAUCAA------GGGCGCGA--AGGCCACAA---UAGU--UGGCUUAA--UGCUCG-G--------CU---ACCCAACUGG--GACUGUG-CCGCU- ....................((.....------(((((...--((((((...---....--))))))..--)))))(-(--------(.---.(((....))--).....)-)))).- ( -22.90) >DroEre_CAF1 230835 110 - 1 AUAUGUUUCCAGUGCUCCUGGCAUUACAAUGCGUGCCGCGAAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA--------CUGACCCCAAACUGGGCGACUGUGGCCGCUU ...(((..((((.....))))....)))..(((.(((((....((((((((........))))))))..(((((((.(.--------.((....))..).)))).))))))))))).. ( -38.60) >DroYak_CAF1 236243 110 - 1 UUUUGUUUACAGUGCUCUUGGCAUUACAAUGCGUGCCGCGAAAGGGCCACAAAUUUAGCUGCGGCCUAAAGUGCUCCGA--------CUGACCCCAAACUGGGCGACUGUGGCCGCUU ..((((....(((((.....))))))))).(((.(((((....(((((.((........)).)))))..(((((((.(.--------.((....))..).)))).))))))))))).. ( -34.50) >DroAna_CAF1 238207 97 - 1 --------------CGGCCGGCAUUGUAAUGCGUGCCGCGAAAGGGCCACAAAUUUAGUUGCGGCCCGGAGAGUAGCUACUAGCUAGCCGACCCCAAACAGGG-------GGCCGCUU --------------.(((((((((........))))).(....)))))............(((((.(((....((((.....)))).))).((((.....)))-------)))))).. ( -41.50) >consensus AUUUGUUU_CAGUGCUCUUGGCAUUACAAUGCGUGCCGCGAAAGGGCCACAAAUUUAGCUGUGGCCUAAAGUGCUCCGA________CUGACCCCAAACUGGGCGACUGUGGCCGCUU ..............................(((.(((((....((((((((........)))))))).........................(((.....))).....)))))))).. (-22.59 = -23.90 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:44 2006