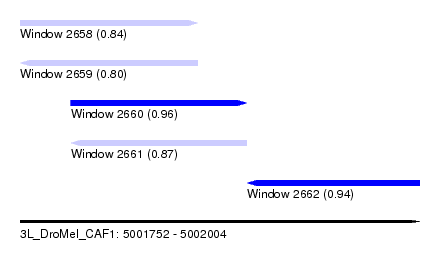
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 5,001,752 – 5,002,004 |
| Length | 252 |
| Max. P | 0.955039 |

| Location | 5,001,752 – 5,001,864 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -31.06 |
| Energy contribution | -31.45 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5001752 112 + 23771897 CAAGUGUU--------UCGGUUAGUGCAGCUGGAGUCUGCUUAGCAUCUUUUUUUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUU .(((((((--------((((((.....)))))))).).))))((((.........(((((((.(..((((((((((........)))))))))).....)))))))).........)))) ( -32.87) >DroSec_CAF1 222363 111 + 1 CGAGUGUU--------UCGGUUAGUGCAGCUGGAGUCUGCUUAGCAUCUUUU-CUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUU ((((...)--------)))(((((.(((((....).))))))))).......-..(((((((.(..((((((((((........)))))))))).....))))))))............. ( -33.20) >DroSim_CAF1 242139 111 + 1 CGAGUGUU--------UCGGUUAGUGCAGCUGGAGUCUGCUUAGCAUCUUUU-UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUU ((((...)--------)))(((((.(((((....).))))))))).......-..(((((((.(..((((((((((........)))))))))).....))))))))............. ( -33.20) >DroEre_CAF1 225498 110 + 1 CGAGUGUU--------UCGGUUAGUGCAGCUGGAGUCUGCUUAGCAUCUUU--UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUU ((((...)--------)))(((((.(((((....).)))))))))......--..(((((((.(..((((((((((........)))))))))).....))))))))............. ( -33.20) >DroYak_CAF1 230468 111 + 1 CGAGUGUU--------UCGGUUAGUGCAGCUGGAGUCUGCUUAGCAUCUUUU-UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUU ((((...)--------)))(((((.(((((....).))))))))).......-..(((((((.(..((((((((((........)))))))))).....))))))))............. ( -33.20) >DroAna_CAF1 232501 118 + 1 CGAGUGUUUUCGGUGUUCGGUGAGUGCAGCUGGAGUCUGUGCAGCAUCUUU--UUUGGCGCCGCGUCGAGCUGCAAUUUCCAAUUUGCAAUUUGCCCGAGGGCGCCAUAAUUAUUUUGUU .(((....)))(((((((((..(((((((.(((((....((((((......--.((((((...))))))))))))..)))))..)))).)))..))...))))))).............. ( -38.11) >consensus CGAGUGUU________UCGGUUAGUGCAGCUGGAGUCUGCUUAGCAUCUUUU_UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUU ...................(((((.(((((....).)))))))))..........(((((((.(..((((((((((........)))))))))).....))))))))............. (-31.06 = -31.45 + 0.39)


| Location | 5,001,752 – 5,001,864 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -24.11 |
| Energy contribution | -24.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5001752 112 - 23771897 AACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAAAAAAAGAUGCUAAGCAGACUCCAGCUGCACUAACCGA--------AACACUUG .............(((((((((.(((....(((((....(....))))))))).).).)))))))...............((((.(....))))).........--------........ ( -26.20) >DroSec_CAF1 222363 111 - 1 AACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAG-AAAAGAUGCUAAGCAGACUCCAGCUGCACUAACCGA--------AACACUCG .............(((((((((.(((....(((((....(....))))))))).).).)))))))..-............((((.(....))))).........--------........ ( -26.20) >DroSim_CAF1 242139 111 - 1 AACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA-AAAAGAUGCUAAGCAGACUCCAGCUGCACUAACCGA--------AACACUCG .............(((((((((.(((....(((((....(....))))))))).).).)))))))..-............((((.(....))))).........--------........ ( -26.20) >DroEre_CAF1 225498 110 - 1 AACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA--AAAGAUGCUAAGCAGACUCCAGCUGCACUAACCGA--------AACACUCG .............(((((((((.(((....(((((....(....))))))))).).).)))))))..--...........((((.(....))))).........--------........ ( -26.20) >DroYak_CAF1 230468 111 - 1 AACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA-AAAAGAUGCUAAGCAGACUCCAGCUGCACUAACCGA--------AACACUCG .............(((((((((.(((....(((((....(....))))))))).).).)))))))..-............((((.(....))))).........--------........ ( -26.20) >DroAna_CAF1 232501 118 - 1 AACAAAAUAAUUAUGGCGCCCUCGGGCAAAUUGCAAAUUGGAAAUUGCAGCUCGACGCGGCGCCAAA--AAAGAUGCUGCACAGACUCCAGCUGCACUCACCGAACACCGAAAACACUCG .............((((((((((((((....(((((........))))))))))).).)))))))..--...(((((.((..........)).))).)).........(((......))) ( -32.50) >consensus AACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA_AAAAGAUGCUAAGCAGACUCCAGCUGCACUAACCGA________AACACUCG .............(((((((((.(((....(((((....(....))))))))).).).)))))))...............((((.(....)))))......................... (-24.11 = -24.00 + -0.11)


| Location | 5,001,784 – 5,001,895 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.29 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5001784 111 + 23771897 UUAGCAUCUUUUUUUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUUGCAAUUAAAAGUCUUGACACUGCCAAAG-----GAA---- .........(((((((((((..(.((((((((((((........))))))...(((....)))((.((((.....)))).))..........)))))).)))))))))-----)))---- ( -30.90) >DroSec_CAF1 222395 110 + 1 UUAGCAUCUUUU-CUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUUGCAAUUAAAAGUCUUGACACUGCCAAAG-----GAA---- .........((.-.((((((..(.((((((((((((........))))))...(((....)))((.((((.....)))).))..........)))))).)))))))..-----)).---- ( -29.00) >DroSim_CAF1 242171 110 + 1 UUAGCAUCUUUU-UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUUGCAAUUAAAAGUCUUGACACUGCCAAAG-----GAA---- .........(((-(((((((..(.((((((((((((........))))))...(((....)))((.((((.....)))).))..........)))))).)))))))))-----)).---- ( -30.80) >DroEre_CAF1 225530 109 + 1 UUAGCAUCUUU--UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUUGCAAUUAAAAGUCUUGACACUGCCAAAG-----GAA---- ........(((--(((((((..(.((((((((((((........))))))...(((....)))((.((((.....)))).))..........)))))).)))))))))-----)).---- ( -30.70) >DroYak_CAF1 230500 110 + 1 UUAGCAUCUUUU-UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUUGCAAUUAAAAGUCUUGACACUGCCAAAG-----GAA---- .........(((-(((((((..(.((((((((((((........))))))...(((....)))((.((((.....)))).))..........)))))).)))))))))-----)).---- ( -30.80) >DroAna_CAF1 232541 118 + 1 GCAGCAUCUUU--UUUGGCGCCGCGUCGAGCUGCAAUUUCCAAUUUGCAAUUUGCCCGAGGGCGCCAUAAUUAUUUUGUUGCAAUUAAAAGUCUUGACACUGCCAAUGCGAAAGAAAAGC ((((((.....--..(((((((.(.(((.(((((((........)))))....)).))))))))))).........)))))).........((((.....(((....))).))))..... ( -33.49) >consensus UUAGCAUCUUUU_UUUGGCGCCGCGUCGAGUUGCAGUUUCCAAUUUGCAAUUUGCCCCAGGGCGCCAUAAUUAUUUUGUUGCAAUUAAAAGUCUUGACACUGCCAAAG_____GAA____ .(((((.........(((((((.(..((((((((((........)))))))))).....)))))))).........)))))....................................... (-27.40 = -27.29 + -0.11)



| Location | 5,001,784 – 5,001,895 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -25.94 |
| Energy contribution | -25.67 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5001784 111 - 23771897 ----UUC-----CUUUGGCAGUGUCAAGACUUUUAAUUGCAACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAAAAAAAGAUGCUAA ----...-----.......((((((........((((((........))))))(((((((((.(((....(((((....(....))))))))).).).))))))).......)))))).. ( -26.90) >DroSec_CAF1 222395 110 - 1 ----UUC-----CUUUGGCAGUGUCAAGACUUUUAAUUGCAACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAG-AAAAGAUGCUAA ----...-----..((((((...((........((((((........))))))(((((((((.(((....(((((....(....))))))))).).).))))))).)-).....)))))) ( -27.40) >DroSim_CAF1 242171 110 - 1 ----UUC-----CUUUGGCAGUGUCAAGACUUUUAAUUGCAACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA-AAAAGAUGCUAA ----...-----.......((((((........((((((........))))))(((((((((.(((....(((((....(....))))))))).).).)))))))..-....)))))).. ( -26.90) >DroEre_CAF1 225530 109 - 1 ----UUC-----CUUUGGCAGUGUCAAGACUUUUAAUUGCAACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA--AAAGAUGCUAA ----...-----.......((((((........((((((........))))))(((((((((.(((....(((((....(....))))))))).).).)))))))..--...)))))).. ( -26.90) >DroYak_CAF1 230500 110 - 1 ----UUC-----CUUUGGCAGUGUCAAGACUUUUAAUUGCAACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA-AAAAGAUGCUAA ----...-----.......((((((........((((((........))))))(((((((((.(((....(((((....(....))))))))).).).)))))))..-....)))))).. ( -26.90) >DroAna_CAF1 232541 118 - 1 GCUUUUCUUUCGCAUUGGCAGUGUCAAGACUUUUAAUUGCAACAAAAUAAUUAUGGCGCCCUCGGGCAAAUUGCAAAUUGGAAAUUGCAGCUCGACGCGGCGCCAAA--AAAGAUGCUGC ((.........))....((((((((........((((((........))))))((((((((((((((....(((((........))))))))))).).)))))))..--...)))))))) ( -38.20) >consensus ____UUC_____CUUUGGCAGUGUCAAGACUUUUAAUUGCAACAAAAUAAUUAUGGCGCCCUGGGGCAAAUUGCAAAUUGGAAACUGCAACUCGACGCGGCGCCAAA_AAAAGAUGCUAA ...................((((((........((((((........))))))(((((((((.(((....(((((....(....))))))))).).).))))))).......)))))).. (-25.94 = -25.67 + -0.28)



| Location | 5,001,895 – 5,002,004 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 5001895 109 - 23771897 AAAAGUUUGCCCUAGAAUCGCAAUUGCUUGGGCGGCGUAAUUGUUCUUUCGCAGCAUUUUC-CCUCCUGGCAAACAAACCGUAAAGGCUGGGGCAUAAAUUGGGCAAAUU ...(((((((((..(((..((((((((.........))))))))...)))...........-..(((..((..((.....))....))..)))........))))))))) ( -28.50) >DroSec_CAF1 222505 110 - 1 AAAAGUUUGCCCUCGAAUUGCAAUUGCUUGGGCGGCGUAAUUGUUCUUUCGCAGCUUUUUCCCCUCUUGGCAAACAAACCGUAGAAGCUGGGGCAUAAAUUGGGCAAAUU ...(((((((((.((((..((((((((.........))))))))...))))((((((((.......(((.....))).....))))))))...........))))))))) ( -31.60) >DroSim_CAF1 242281 109 - 1 AAAAGUUUGCCCUCGAAUUGCAAUUGCUUGGGCGGCGUAAUUGUUCUUUCGCAGCUUUUUC-CCUCUUGGCAAACAAACCGUAGAAGCUGGGGCAUAAAUUGGGCAAAUU ...(((((((((.((((..((((((((.........))))))))...))))((((((((..-....(((.....))).....))))))))...........))))))))) ( -32.20) >DroEre_CAF1 225639 108 - 1 AAAAGUUUGCCCUCGAAUUGCAAUUGCUUGGGCGGCGUAAUUGUUCUUUCGCAGCUUUUUC--CUCUUGGCAAACAAACCGUAAAAGCUGGGGCAUAAAUUGGGCAAAUU ...(((((((((.((((..((((((((.........))))))))...))))((((((((..--...(((.....))).....))))))))...........))))))))) ( -32.70) >DroYak_CAF1 230610 109 - 1 AAAAGUUUGCCCUCGAAUUGCAAUUGCUUGGGCGGCGUAAUUGUUCUUUCGCAGCUUCUUC-UCACUUGGCAAGCAAACCGUAAAAGCUGGGGCAUAAAUUGGGCAAAUU ...(((((((((......(((....((((..((((......(((......)))((((((..-......)).))))...))))..))))....)))......))))))))) ( -32.90) >consensus AAAAGUUUGCCCUCGAAUUGCAAUUGCUUGGGCGGCGUAAUUGUUCUUUCGCAGCUUUUUC_CCUCUUGGCAAACAAACCGUAAAAGCUGGGGCAUAAAUUGGGCAAAUU ...(((((((((......(((....((((..((((......(((......)))(((............))).......))))..))))....)))......))))))))) (-28.70 = -28.94 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:38 2006