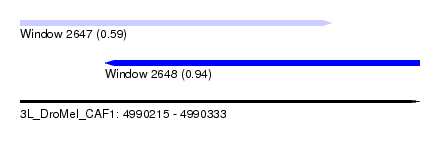
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,990,215 – 4,990,333 |
| Length | 118 |
| Max. P | 0.939839 |

| Location | 4,990,215 – 4,990,307 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -13.82 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4990215 92 + 23771897 GCCCCG--GCUCACAGCGAUAAAUCACAACGAAAUUCACAUAAUUUUUAUCGUUUUGCACAAUUUUGCAUGUGCUCACUUACUACAUACAUAA----A ((....--))....(((((((((......................)))))))))..(((((........)))))...................----. ( -13.35) >DroSec_CAF1 210800 96 + 1 GCCCCG--GCUCACAGCGAUAAAUCACAACGAAAUUCACAUAAUUUUUAUCGUUUUGCACAAUUUUGCAUGUGCUCACUUACUACAUACAUAUAUGUA ((....--))....(((((((((......................)))))))))..(((((........)))))........((((((....)))))) ( -15.45) >DroSim_CAF1 230544 96 + 1 GCCCCG--GCUCACAGCGAUAAAUCACAACGAAAUUCACAUAAUUUUUAUCGUUUUGCACAAUUUUGCAUGUGCUCACUUACUACAUACAUAUAUGUA ((....--))....(((((((((......................)))))))))..(((((........)))))........((((((....)))))) ( -15.45) >DroYak_CAF1 218718 93 + 1 GCCCCAACACUCACAGCGAUAAAUCACAACGAAAUUCACAUAAUUUUUAUCGUUUUGCACAAUUUUGCAUGUGCCCACUUACUACAUAUAUC-----U ..............(((((((((......................)))))))))..(((((........)))))..................-----. ( -11.05) >consensus GCCCCG__GCUCACAGCGAUAAAUCACAACGAAAUUCACAUAAUUUUUAUCGUUUUGCACAAUUUUGCAUGUGCUCACUUACUACAUACAUAU____A ..............(((((((((......................)))))))))..(((((........)))))........................ (-11.35 = -11.35 + -0.00)

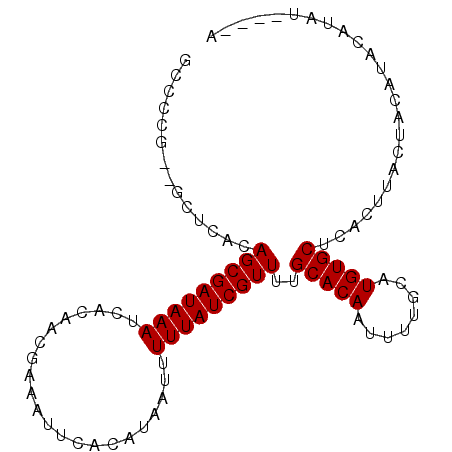
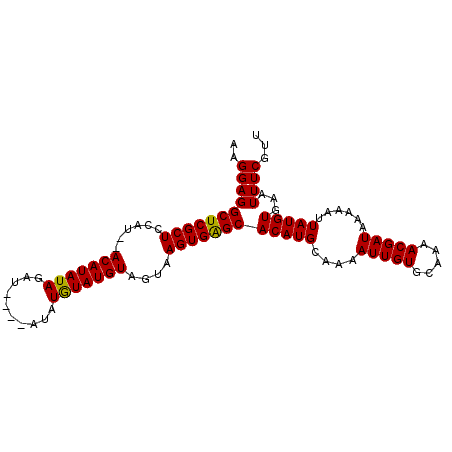
| Location | 4,990,240 – 4,990,333 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -20.17 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4990240 93 - 23771897 AAGGAGGCUCGCUCCAU--ACAUAUAGAU----UUAUGUAUGUAGUAAGUGAGCACAUGCAAAAUUGUGCAAAACGAUAAAAAUUAUGUGAAUUUCGUU ..(((((((((((...(--(((((((...----...))))))))...)))))))(((((....(((((.....)))))......)))))...))))... ( -24.50) >DroSec_CAF1 210825 97 - 1 AAGGAGGCUCGCUCCAU--ACAUAUAGAUACAUAUAUGUAUGUAGUAAGUGAGCACAUGCAAAAUUGUGCAAAACGAUAAAAAUUAUGUGAAUUUCGUU ......(((..((.(((--(((((((......)))))))))).))..)))..(((((........)))))..(((((.................))))) ( -25.93) >DroSim_CAF1 230569 97 - 1 AAGGAGGCUCGCUCCAU--ACAUAUAGAUACAUAUAUGUAUGUAGUAAGUGAGCACAUGCAAAAUUGUGCAAAACGAUAAAAAUUAUGUGAAUUUCGUU ......(((..((.(((--(((((((......)))))))))).))..)))..(((((........)))))..(((((.................))))) ( -25.93) >DroYak_CAF1 218745 94 - 1 AAGGAGGCUCGCUCCAUACACAUAUAGAA-----GAUAUAUGUAGUAAGUGGGCACAUGCAAAAUUGUGCAAAACGAUAAAAAUUAUGUGAAUUUCGUU ...((((.((((.(((((((((((((...-----..))))))).))..))))(((((........))))).................)))).))))... ( -22.90) >consensus AAGGAGGCUCGCUCCAU__ACAUAUAGAU____AUAUGUAUGUAGUAAGUGAGCACAUGCAAAAUUGUGCAAAACGAUAAAAAUUAUGUGAAUUUCGUU ..(((((((((((......(((((((..........)))))))....)))))))(((((....(((((.....)))))......)))))...))))... (-20.17 = -19.80 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:24 2006