| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 527,520 – 527,736 |
| Length | 216 |
| Max. P | 0.999568 |

| Location | 527,520 – 527,636 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 527520 116 + 23771897 AUGUAGUCCGUAAAGUGGGUAAGUCUGGAAUGCCCUGGAACUAAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUU .....(((((.(((((((..((((.((....((((((..(((.....)))..)))..)))....))))))..))))))).((((......)))).......))))).......... ( -32.30) >DroSec_CAF1 1771 99 + 1 AUAUU-----------------GUCUGGAAUGCCCUGGAACAAAAUAAGUCUCAGUAGGUUGAAAAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUU .....-----------------........(((((.((((......(((((((((.((((.((........)).)))))))))))))........))))...)))))......... ( -24.34) >DroSim_CAF1 1764 99 + 1 AUAUU-----------------GUCUGGAAAGCCCUGGAACAGAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUU .....-----------------.........((((.((((..((..(((((((((.((((.((........)).)))))))))))))...))...))))...)))).......... ( -24.50) >DroEre_CAF1 1753 116 + 1 UCCUUCUCCAUGAAGUGUGUUACUCUAGAAGCUUCUACAACAAAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUCUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUGUAUU ...........(((((...((......)).)))))....(((((..(((((((((.((((.((........)).))))))))))))).....(((.......)))...)))))... ( -23.70) >DroYak_CAF1 1845 116 + 1 UCGUUUUCCAUGAAGUGUGUUGGUCUAGAAUGCUCUGCAACAGAAUGAGUCCCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUU ..(((.(((..((((..(((((...((((....)))))))))((..(((((((((.((((.((........)).)))))))))))))...))..))))....))).)))....... ( -24.60) >consensus AUAUU_UCC_U_AAGUG_GU__GUCUGGAAUGCCCUGGAACAAAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUU ...............................((((.((((......(((((((((.((((.((........)).)))))))))))))........))))...)))).......... (-19.56 = -19.48 + -0.08)


| Location | 527,520 – 527,636 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -18.14 |
| Energy contribution | -19.18 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 527520 116 - 23771897 AAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUUAGUUCCAGGGCAUUCCAGACUUACCCACUUUACGGACUACAU ...........(((.......)))......(((((..(((((((.................(((..(((.....)))..)))((....))........)))))))..))))).... ( -26.80) >DroSec_CAF1 1771 99 - 1 AAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUUUUCAACCUACUGAGACUUAUUUUGUUCCAGGGCAUUCCAGAC-----------------AAUAU .........(((((...((((.(((((((..(((((....).)))).....(((((......))))).))))))).)))).)))))........-----------------..... ( -27.20) >DroSim_CAF1 1764 99 - 1 AAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUCUGUUCCAGGGCUUUCCAGAC-----------------AAUAU ..........((((...((((.(((((((..(((((....).))))......((((......))))..))))))).)))).)))).........-----------------..... ( -27.70) >DroEre_CAF1 1753 116 - 1 AAUACAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAGAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUUUGUUGUAGAAGCUUCUAGAGUAACACACUUCAUGGAGAAGGA ............(((.......(((((....))))).((((((((((((.((((((......))))...)).)))))).((((....))))........)))))).)))....... ( -23.00) >DroYak_CAF1 1845 116 - 1 AAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGGGACUCAUUCUGUUGCAGAGCAUUCUAGACCAACACACUUCAUGGAAAACGA ............(((...(((((((((...(((((((...((((..............))))))))))).)))))((((.((......))....))))...)))).)))....... ( -22.24) >consensus AAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUUUGUUCCAGGGCAUUCCAGAC__AC_CACUU_A_GGA_AACAU ..........((((...((((.(((((((..(((((....).))))......((((......))))..))))))).)))).))))............................... (-18.14 = -19.18 + 1.04)

| Location | 527,560 – 527,663 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 97.09 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -23.04 |
| Energy contribution | -22.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 527560 103 + 23771897 CUAAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUG ...((((((((((((.....((((....((((((......((((......))))........))))))..))))....))))))))))))............. ( -22.84) >DroSec_CAF1 1794 103 + 1 CAAAAUAAGUCUCAGUAGGUUGAAAAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUG ...((((((((((((.....((((((..((((((......((((......))))........))))))))))))....))))))))))))............. ( -23.84) >DroSim_CAF1 1787 103 + 1 CAGAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUG ...((((((((((((.....((((....((((((......((((......))))........))))))..))))....))))))))))))............. ( -22.94) >DroEre_CAF1 1793 103 + 1 CAAAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUCUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUGUAUUGUUUGGGAUUUAUUGCCAACUCUUUUG (((((.(((((((((.((((.((........)).)))))))))))))............((((((((((....))))))))))...............))))) ( -24.30) >DroYak_CAF1 1885 103 + 1 CAGAAUGAGUCCCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUG ...((((((((((((.....((((....((((((......((((......))))........))))))..))))....))))))))))))............. ( -26.04) >consensus CAAAAUAAGUCUCAGUAGGUUGAAUAAUUUGUCCACUUUUGGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUG ...((((((((((((.....((((....((((((......((((......))))........))))))..))))....))))))))))))............. (-23.04 = -22.92 + -0.12)

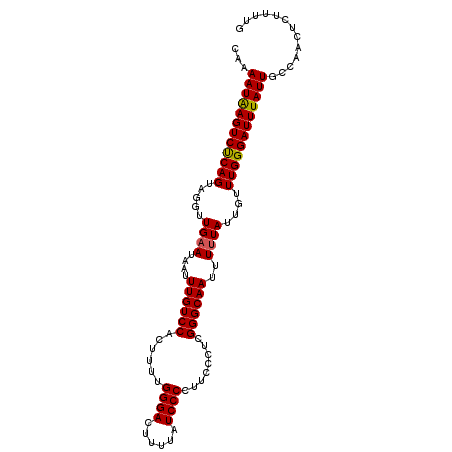
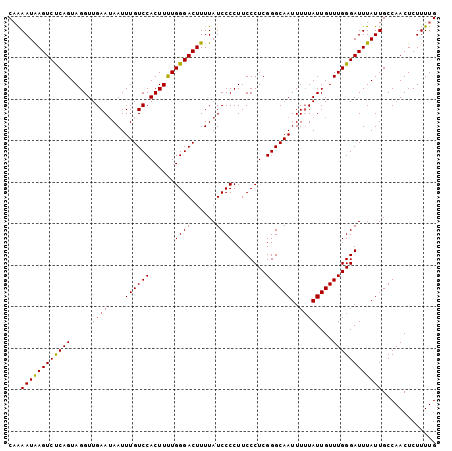
| Location | 527,560 – 527,663 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 97.09 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 527560 103 - 23771897 CAAAAGAGUUGGCAAUAAAUCCCAAACAAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUUAG .(((.(((((((((((..................))))))..(((.....((((((..(((((....).))))...))))))..))).....))))).))).. ( -19.27) >DroSec_CAF1 1794 103 - 1 CAAAAGAGUUGGCAAUAAAUCCCAAACAAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUUUUCAACCUACUGAGACUUAUUUUG (((((..(((.((......((((...((((....)))).....))))..(((((....)))))....)).))).....(((((......)))))....))))) ( -20.40) >DroSim_CAF1 1787 103 - 1 CAAAAGAGUUGGCAAUAAAUCCCAAACAAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUCUG ....((((((.((......((((...((((....)))).....))))..(((((....)))))....)).)))......((((......))))......))). ( -19.60) >DroEre_CAF1 1793 103 - 1 CAAAAGAGUUGGCAAUAAAUCCCAAACAAUACAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAGAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUUUG ((((((((((((((((..................))))))..(((.....((((((..(((((....).))))...))))))..))).....))))).))))) ( -20.27) >DroYak_CAF1 1885 103 - 1 CAAAAGAGUUGGCAAUAAAUCCCAAACAAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGGGACUCAUUCUG ....(((((.........(((((...((((....))))((....))...)))))...(((((((...((((..............))))))))))).))))). ( -23.34) >consensus CAAAAGAGUUGGCAAUAAAUCCCAAACAAUAAAAAUUGCCCGAGGGAAGGGGAUAAAAGUCCCAAAAGUGGACAAAUUAUUCAACCUACUGAGACUUAUUUUG ((..((.(((((......(((((...((((....))))((....))...)))))....(((((....).)))).......)))))))..))............ (-19.24 = -19.24 + 0.00)

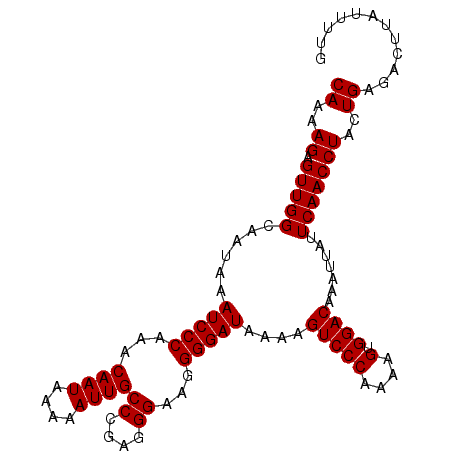
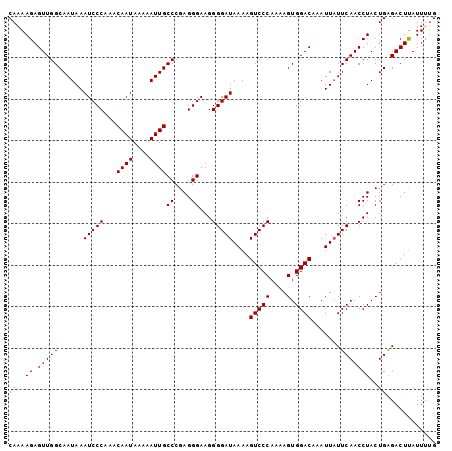
| Location | 527,600 – 527,696 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.82 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.12 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 527600 96 + 23771897 GGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACAAUAGCUG ((((......)))).....((((((((((....))))))))))..((((((....((((((((.............))))))))...))))))... ( -25.32) >DroSec_CAF1 1834 95 + 1 GGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACGAU-GCUG ((((......)))).....((((((((((....))))))))))...(((((....((((((((.............))))))))...))))-)... ( -24.12) >DroSim_CAF1 1827 96 + 1 GGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACGAUAGCUG ((((......)))).....((((((((((....))))))))))..((((((....((((((((.............))))))))...))))))... ( -25.02) >DroEre_CAF1 1833 96 + 1 GGGACUUUUAUCCCCUUCCCUCGGGCAAUUUGUAUUGUUUGGGAUUUAUUGCCAACUCUUUUGGCGUUUAUCAAAACAAAAGAGGCACGAUAACUG ((((......)))).....((((((((((....))))))))))..((((((....((((((((.............))))))))...))))))... ( -25.32) >DroYak_CAF1 1925 96 + 1 GGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGCACGAUAACUG ((((......)))).....((((((((((....))))))))))..((((((....((((((((.............))))))))...))))))... ( -25.32) >consensus GGGACUUUUAUCCCCUUCCCUCGGGCAAUUUUUAUUGUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACGAUAGCUG ((((......)))).....((((((((((....))))))))))..((((((....((((((((.............))))))))...))))))... (-25.02 = -24.82 + -0.20)

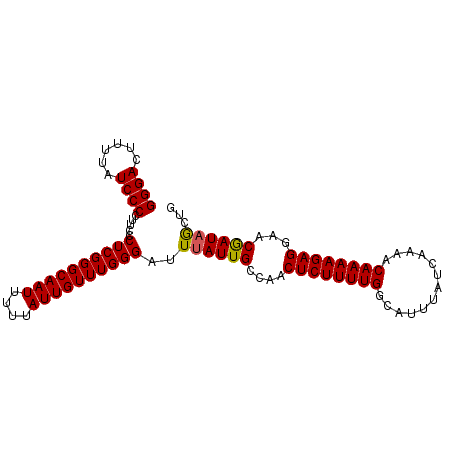
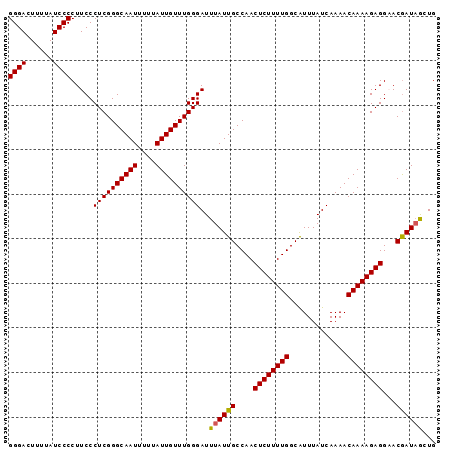
| Location | 527,636 – 527,736 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -24.88 |
| Energy contribution | -26.42 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 527636 100 + 23771897 GUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACAAUAGCUGGCAGGCGGAAAUGGAAAUAACUGCCUUAUCCCUUCUCUCC ....(((((....(((((.((((((((.............))))))))..........)))))(((((..((....))..)))))..)))))........ ( -27.62) >DroSec_CAF1 1870 99 + 1 GUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACGAU-GCUGGCAGGCGGAAAUGGAAAUAACUGCCUUAUCCCUUCUCUCC ....(((((....(((((.((((((((.............)))))))).......-..)))))(((((..((....))..)))))..)))))........ ( -27.72) >DroSim_CAF1 1863 100 + 1 GUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACGAUAGCUGGCAGGCGGAAAUGGAAAUAACUGCUUUAUCCCUUCUCUCC ((((..(((....((((((.....))))))...))).))))...((((((..(((((..(((((................))))))))))..)))))).. ( -25.59) >DroEre_CAF1 1869 100 + 1 GUUUGGGAUUUAUUGCCAACUCUUUUGGCGUUUAUCAAAACAAAAGAGGCACGAUAACUGGCUGGCGGAAAUGGAAAUAACUGCCUUAUCCCUCCUCCUA ....(((((.....((((.((((((((.............))))))))..........)))).(((((..((....))..)))))..)))))........ ( -26.02) >DroYak_CAF1 1961 100 + 1 GUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGCACGAUAACUGGCAGGCGGAAAUGGAAAUAACUGCCUUAUCCCUCCUCUCC ((((..(((....((((((.....))))))...))).))))...(((((...(((((..(((((................))))))))))...))))).. ( -28.19) >consensus GUUUGGGAUUUAUUGCCAACUCUUUUGGCAUUUAUCAAAACAAAAGAGGAACGAUAGCUGGCAGGCGGAAAUGGAAAUAACUGCCUUAUCCCUUCUCUCC ....(((((....(((((.((((((((.............))))))))..........)))))(((((..((....))..)))))..)))))........ (-24.88 = -26.42 + 1.54)

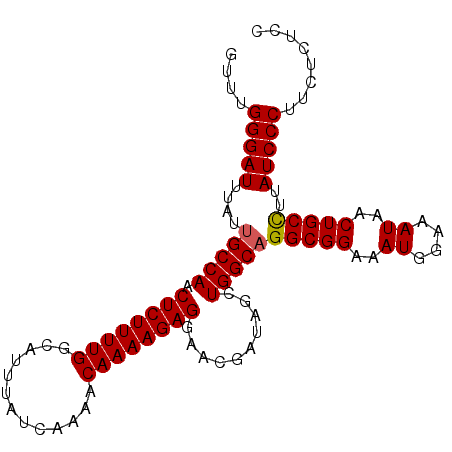
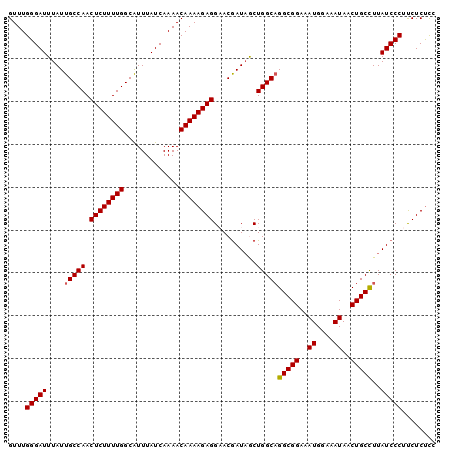
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:23 2006