
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,979,925 – 4,980,026 |
| Length | 101 |
| Max. P | 0.993233 |

| Location | 4,979,925 – 4,980,026 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.87 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -9.93 |
| Energy contribution | -11.90 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
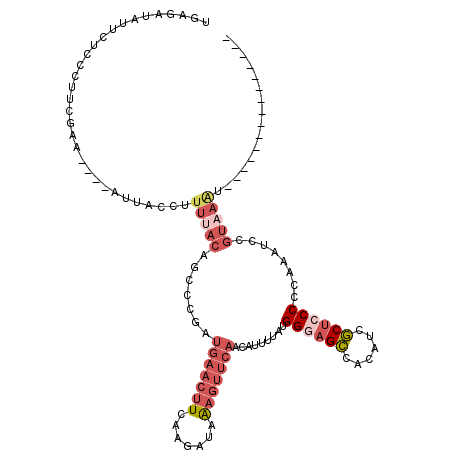
>3L_DroMel_CAF1 4979925 101 + 23771897 UGAGAUAUUCUCCCUUCGAA----AUUACCUUUUACAGCCCGAUGAACUUCAAGAUAAAGUUCAACAUUUUAUGGGAGCCACAUCGCUCCCCCAAAUCCGUAAAU--------------- .(((.....)))........----.......(((((.....(.(((((((.......))))))).).......((((((......))))))........))))).--------------- ( -21.40) >DroPse_CAF1 255408 111 + 1 AAACUCUUUCCCCGCCCGAACUAGACUUCCCUCAAAGAUCCCAUAAA--UCCAUAAAGAAGCCAAUAUU-CGGGGCAGG--CAUUUCUCCCCCUG----GAAAGUCUCACCUUUGUCUGG ...........(((..((((..((((((.((................--........(((.......))-)((((.(((--....))).)))).)----).))))))....))))..))) ( -21.00) >DroSec_CAF1 200595 101 + 1 UGGGAUAUUCUCCCUUCGAA----AUUACAUUUUACAGCCCGAUGAACUUCAAGAUAAAGUUCAACAUUUUAUGGGAGCCACAUCGCUCCCCCAAAUCCGUAAAU--------------- .((((.....))))......----.......(((((.....(.(((((((.......))))))).).......((((((......))))))........))))).--------------- ( -24.50) >DroSim_CAF1 220018 101 + 1 UGAGAUAUUCUCCCUUCGAA----AUUACAUUUUACAGCCCGAUGAACUUCAAGAUAAAGUUCAACAUUUUAUGGGAGCCACAUCGCUCCCCCAAAUCCGUAAAU--------------- .(((.....)))........----.......(((((.....(.(((((((.......))))))).).......((((((......))))))........))))).--------------- ( -21.40) >DroEre_CAF1 203764 81 + 1 -GAGAUAUU-----------------------UUACAGCCCGACGAACUUCAAGAUAAAGUUCAACAUUUUAUGGGUGCCACAUCGCUCCCCCAAAUCCGUAAAU--------------- -.......(-----------------------((((........((((((.......))))))..........(((.((......)).)))........))))).--------------- ( -14.40) >DroYak_CAF1 207857 101 + 1 UGAGAUAUUCUCCCCUCGAA----AAUUCCUUUAACAGCCCGAUGAACUUCAAGAUAAAGUUCAACAUUUUAUGGCAGCCACAUUGCUCCCCCAAAUCCGUAAAU--------------- .(((.....)))........----.................(.(((((((.......))))))).)..(((((((.(((......))).........))))))).--------------- ( -11.80) >consensus UGAGAUAUUCUCCCUUCGAA____AUUACCUUUUACAGCCCGAUGAACUUCAAGAUAAAGUUCAACAUUUUAUGGGAGCCACAUCGCUCCCCCAAAUCCGUAAAU_______________ ...............................(((((.......(((((((.......))))))).........((((((......))))))........)))))................ ( -9.93 = -11.90 + 1.97)


| Location | 4,979,925 – 4,980,026 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.87 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -16.17 |
| Energy contribution | -18.37 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4979925 101 - 23771897 ---------------AUUUACGGAUUUGGGGGAGCGAUGUGGCUCCCAUAAAAUGUUGAACUUUAUCUUGAAGUUCAUCGGGCUGUAAAAGGUAAU----UUCGAAGGGAGAAUAUCUCA ---------------.(((((((.(((((((((((......)))))).........((((((((.....))))))))))))))))))))(((((.(----(((.....)))).))))).. ( -32.30) >DroPse_CAF1 255408 111 - 1 CCAGACAAAGGUGAGACUUUC----CAGGGGGAGAAAUG--CCUGCCCCG-AAUAUUGGCUUCUUUAUGGA--UUUAUGGGAUCUUUGAGGGAAGUCUAGUUCGGGCGGGGAAAGAGUUU ................(((((----(..(((.(....).--)))((((.(-(((...(((((((((..(((--((.....)))))...)))))))))..))))))))..))))))..... ( -38.10) >DroSec_CAF1 200595 101 - 1 ---------------AUUUACGGAUUUGGGGGAGCGAUGUGGCUCCCAUAAAAUGUUGAACUUUAUCUUGAAGUUCAUCGGGCUGUAAAAUGUAAU----UUCGAAGGGAGAAUAUCCCA ---------------.(((((((.(((((((((((......)))))).........((((((((.....)))))))))))))))))))).......----......((((.....)))). ( -33.90) >DroSim_CAF1 220018 101 - 1 ---------------AUUUACGGAUUUGGGGGAGCGAUGUGGCUCCCAUAAAAUGUUGAACUUUAUCUUGAAGUUCAUCGGGCUGUAAAAUGUAAU----UUCGAAGGGAGAAUAUCUCA ---------------.(((((((.(((((((((((......)))))).........((((((((.....)))))))))))))))))))).......----......((((.....)))). ( -30.90) >DroEre_CAF1 203764 81 - 1 ---------------AUUUACGGAUUUGGGGGAGCGAUGUGGCACCCAUAAAAUGUUGAACUUUAUCUUGAAGUUCGUCGGGCUGUAA-----------------------AAUAUCUC- ---------------.(((((((.((((((((.((......)).)))..........(((((((.....))))))).)))))))))))-----------------------).......- ( -21.80) >DroYak_CAF1 207857 101 - 1 ---------------AUUUACGGAUUUGGGGGAGCAAUGUGGCUGCCAUAAAAUGUUGAACUUUAUCUUGAAGUUCAUCGGGCUGUUAAAGGAAUU----UUCGAGGGGAGAAUAUCUCA ---------------....((((.(((((((.(((......))).)).........((((((((.....)))))))))))))))))...(((.(((----(((.....)))))).))).. ( -22.80) >consensus _______________AUUUACGGAUUUGGGGGAGCGAUGUGGCUCCCAUAAAAUGUUGAACUUUAUCUUGAAGUUCAUCGGGCUGUAAAAGGUAAU____UUCGAAGGGAGAAUAUCUCA ................(((((((.(((((((((((......)))))).........(((((((((...)))))))))))))))))))))............................... (-16.17 = -18.37 + 2.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:14 2006