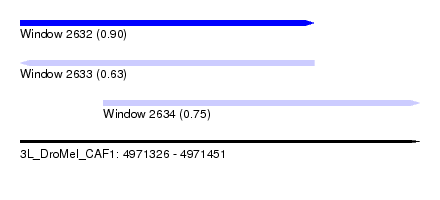
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,971,326 – 4,971,451 |
| Length | 125 |
| Max. P | 0.901287 |

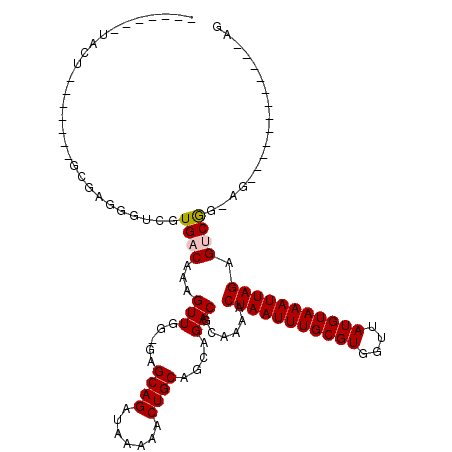
| Location | 4,971,326 – 4,971,418 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4971326 92 + 23771897 -------UACU-------GCGAGGGUCGUGACAAAGUUGG-GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG-AG------------AG -------..((-------.((((.(((....((....)).-..((((.......))))....))).)....(((((((((((....)))))))))))..))).-))------------.. ( -23.90) >DroPse_CAF1 247099 120 + 1 ACCCACAGCCUGCAGGAGGCUGCAGUCGUGACAAAGUUGUGCAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGACAGGAGCGAAACGGACCAAG ..((.((((((.....))))))...((((..(...(((.(((.((((.......)))).))))))......(((((((((((....))))))))))).....)..))))...))...... ( -36.20) >DroEre_CAF1 193100 92 + 1 -------CACU-------GCGAGGGUCGUGACAAAGUUGG-GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGC-AG------------CG -------..((-------(((((.(((....((....)).-..((((.......))))....))).)....(((((((((((....)))))))))))..))))-))------------.. ( -27.70) >DroYak_CAF1 199061 92 + 1 -------UACU-------GCGAGGGUCGUGACAAAGUUGG-GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG-AG------------AG -------..((-------.((((.(((....((....)).-..((((.......))))....))).)....(((((((((((....)))))))))))..))).-))------------.. ( -23.90) >DroAna_CAF1 200681 94 + 1 -------UAGU---UGAGGCGAGGGUCGUGACAAAGUUGG-GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG-A--------------G -------....---...(((..(.(((....((....)).-..((((.......))))....))).)....(((((((((((....))))))))))).)))..-.--------------. ( -21.70) >DroPer_CAF1 231839 120 + 1 ACCCACAGCCUGCAGGAGGCUGCAGUCGUGACAAAGUUGUGCAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGACAGGAGCGAAACGGACCAAG ..((.((((((.....))))))...((((..(...(((.(((.((((.......)))).))))))......(((((((((((....))))))))))).....)..))))...))...... ( -36.20) >consensus _______UACU_______GCGAGGGUCGUGACAAAGUUGG_GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG_AG____________AG ............................((((...(((.....((((.......))))....)))......(((((((((((....))))))))))).)))).................. (-17.74 = -17.85 + 0.11)

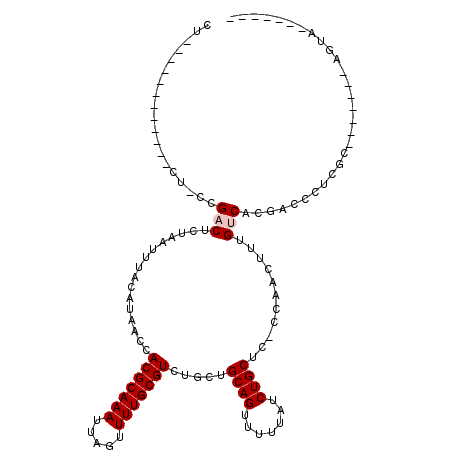
| Location | 4,971,326 – 4,971,418 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -12.93 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4971326 92 - 23771897 CU------------CU-CCGACUCUAAUUUACAUAACCACGCAAAUUAGUUUUGCGUCUGCUGCAGUUUUUAUCUGCUC-CCAACUUUGUCACGACCCUCGC-------AGUA------- ..------------..-.....................(((((((.....)))))))((((.((((.......))))..-........((....))....))-------))..------- ( -15.50) >DroPse_CAF1 247099 120 - 1 CUUGGUCCGUUUCGCUCCUGUCUCUAAUUUACAUAACCACGCAAAUUAGUUUUGCGUCUGCUGCAGUUUUUAUCUGCUGCACAACUUUGUCACGACUGCAGCCUCCUGCAGGCUGUGGGU .......(((..(.........................(((((((.....))))))).(((.((((.......)))).))).......)..))).(..((((((.....))))))..).. ( -31.40) >DroEre_CAF1 193100 92 - 1 CG------------CU-GCGACUCUAAUUUACAUAACCACGCAAAUUAGUUUUGCGUCUGCUGCAGUUUUUAUCUGCUC-CCAACUUUGUCACGACCCUCGC-------AGUG------- ((------------((-((((.................(((((((.....))))))).....((((.......))))..-..................))))-------))))------- ( -23.50) >DroYak_CAF1 199061 92 - 1 CU------------CU-CCGACUCUAAUUUACAUAACCACGCAAAUUAGUUUUGCGUCUGCUGCAGUUUUUAUCUGCUC-CCAACUUUGUCACGACCCUCGC-------AGUA------- ..------------..-.....................(((((((.....)))))))((((.((((.......))))..-........((....))....))-------))..------- ( -15.50) >DroAna_CAF1 200681 94 - 1 C--------------U-CCGACUCUAAUUUACAUAACCACGCAAAUUAGUUUUGCGUCUGCUGCAGUUUUUAUCUGCUC-CCAACUUUGUCACGACCCUCGCCUCA---ACUA------- .--------------.-..(((................(((((((.....))))))).....((((.......))))..-........)))...............---....------- ( -14.90) >DroPer_CAF1 231839 120 - 1 CUUGGUCCGUUUCGCUCCUGUCUCUAAUUUACAUAACCACGCAAAUUAGUUUUGCGUCUGCUGCAGUUUUUAUCUGCUGCACAACUUUGUCACGACUGCAGCCUCCUGCAGGCUGUGGGU .......(((..(.........................(((((((.....))))))).(((.((((.......)))).))).......)..))).(..((((((.....))))))..).. ( -31.40) >consensus CU____________CU_CCGACUCUAAUUUACAUAACCACGCAAAUUAGUUUUGCGUCUGCUGCAGUUUUUAUCUGCUC_CCAACUUUGUCACGACCCUCGC_______AGUA_______ ...................(((................(((((((.....))))))).....((((.......))))...........)))............................. (-12.93 = -13.27 + 0.33)

| Location | 4,971,352 – 4,971,451 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
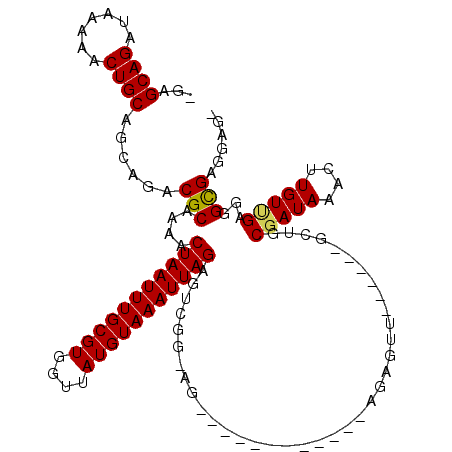
>3L_DroMel_CAF1 4971352 99 + 23771897 -GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG-AG------------AGAGUU------GCUGCGAUAAACUUGUCGAGGGCGAGGAG- -..((((.......)))).((((..((....(((((((((((....)))))))))))..((..-..------------.)))).------.))))......(((((.....)))))...- ( -26.70) >DroPse_CAF1 247139 119 + 1 GCAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGACAGGAGCGAAACGGACCAAGAGUUCCAGCAGCAGCGAUAAACUUGUUGAGGGUGUGAGG- ((.((((.......)))).))..((((....(((((((((((....))))))))))).....(((((..............)))))..(..((((((.....))))))..)))))....- ( -30.74) >DroSec_CAF1 192021 100 + 1 -GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG-AG------------AGAGUU------GCUGCGAUAAACUUGUCGAGGGCGAGGAGC -..((..........((((((((.(......(((((((((((....)))))))))))..((..-..------------.)))))------)))))).....(((((.....)))))..)) ( -26.80) >DroEre_CAF1 193126 99 + 1 -GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGC-AG------------CGAGUU------GGUGCGAUAAACUUGUGGAGGGCGAAGAG- -..((((.......)))).((...(((((..(((((((((((....))))))))))).(((((-(.------------(.....------).))))))....)))))....))......- ( -30.60) >DroAna_CAF1 200711 93 + 1 -GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG-A--------------GAGCU------GCUGCGAUAAACUUGUUGAGGGCGA----- -..((.(((((....((((((((...(....(((((((((((....))))))))))).....)-.--------------...))------))))))......)))))....))..----- ( -25.60) >DroPer_CAF1 231879 119 + 1 GCAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGACAGGAGCGAAACGGACCAAGAGUUCCAGCAGCAGCGAUAAACUUGUUGAGGGUGUGAGG- ((.((((.......)))).))..((((....(((((((((((....))))))))))).....(((((..............)))))..(..((((((.....))))))..)))))....- ( -30.74) >consensus _GAGCAGAUAAAAACUGCAGCAGACGCAAAACUAAUUUGCGUGGUUAUGUAAAUUAGAGUCGG_AG____________AGAGUU______GCUGCGAUAAACUUGUUGAGGGCGAGGAG_ ...((((.......))))......(((....(((((((((((....))))))))))).....................................(((((....)))))...)))...... (-19.67 = -19.45 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:10 2006