| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
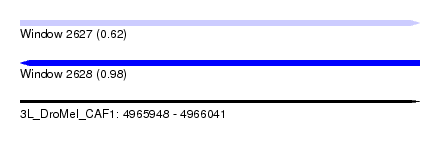
| Location | 4,965,948 – 4,966,041 |
| Length | 93 |
| Max. P | 0.975900 |

| Location | 4,965,948 – 4,966,041 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4965948 93 + 23771897 CACAAAC--------AAACACACAAACGCCCACCAAAAAGGAUACCCAUCAAUGAGU-GUUUUGUGUGUGUUGUGUGU-------UGUUUGUUG-------UUCGUUAUUUGUUGU .((((((--------((.(((((((.(((.(((.....(((((((.((....)).))-)))))))).)))))))))))-------)))))))..-------............... ( -26.30) >DroSec_CAF1 186677 100 + 1 CACAAAC--------AAACACACAAACGCCCACCAAAAAGGAUACCCAUCAAUGAGU-GUUUUGUGUGUGUUGUGUGUUGUGCGUUGUUUGUUG-------CUCGUUAUUUGUUGU .((((((--------((.(.(((((((((.(((((...(((((((.((....)).))-))))).)).)))..)))).))))).)))))))))..-------............... ( -25.80) >DroSim_CAF1 205894 100 + 1 CACAAAC--------AAACACACAAACGCCCACCAAAAAGGAUACCCAUCAAUGAGU-GUUUUGUGUGUGUUGUGUGUUGUGCGUUGUUUGUUG-------UUCGUUAUUUGUUGU .((((((--------((.(.(((((((((.(((((...(((((((.((....)).))-))))).)).)))..)))).))))).)))))))))..-------............... ( -25.80) >DroYak_CAF1 193484 116 + 1 CACAAACACAAAUACAAACACACGGCCGCCCACCAAAAAGGAUACCCAUCAAUGAGUGGUAUUGUGUGCGUUGUGUGGUGUGUGUUGUUUGUUGUUCGUUGUACGUUAUUUGUUGU .((((..((((..((((.((((((.(((((.((((...(.(((((((........).)))))).).)).)).).))))))))))))))...))))...)))).............. ( -31.20) >consensus CACAAAC________AAACACACAAACGCCCACCAAAAAGGAUACCCAUCAAUGAGU_GUUUUGUGUGUGUUGUGUGUUGUGCGUUGUUUGUUG_______UUCGUUAUUUGUUGU .((((((.........(((((((((.(((.(((.((((.....((.((....)).))..))))))).)))))))))))).......))))))........................ (-19.93 = -20.12 + 0.19)


| Location | 4,965,948 – 4,966,041 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.85 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4965948 93 - 23771897 ACAACAAAUAACGAA-------CAACAAACA-------ACACACAACACACACAAAAC-ACUCAUUGAUGGGUAUCCUUUUUGGUGGGCGUUUGUGUGUUU--------GUUUGUG ...............-------..(((((((-------(........(((((((((.(-.(((((..(.(((...)))..)..))))).))))))))))))--------)))))). ( -26.60) >DroSec_CAF1 186677 100 - 1 ACAACAAAUAACGAG-------CAACAAACAACGCACAACACACAACACACACAAAAC-ACUCAUUGAUGGGUAUCCUUUUUGGUGGGCGUUUGUGUGUUU--------GUUUGUG ..............(-------(..........))...(((.((((.(((((((((.(-.(((((..(.(((...)))..)..))))).))))))))))))--------)).))). ( -26.20) >DroSim_CAF1 205894 100 - 1 ACAACAAAUAACGAA-------CAACAAACAACGCACAACACACAACACACACAAAAC-ACUCAUUGAUGGGUAUCCUUUUUGGUGGGCGUUUGUGUGUUU--------GUUUGUG ...............-------............(((((.(((.((((((((...(((-.(((((..(.(((...)))..)..))))).))))))))))))--------))))))) ( -26.10) >DroYak_CAF1 193484 116 - 1 ACAACAAAUAACGUACAACGAACAACAAACAACACACACCACACAACGCACACAAUACCACUCAUUGAUGGGUAUCCUUUUUGGUGGGCGGCCGUGUGUUUGUAUUUGUGUUUGUG ...........((.....))........((((.(((((....((((.((((((....((.(((((..(.(((...)))..)..))))).))..))))))))))...))))))))). ( -28.40) >consensus ACAACAAAUAACGAA_______CAACAAACAACGCACAACACACAACACACACAAAAC_ACUCAUUGAUGGGUAUCCUUUUUGGUGGGCGUUUGUGUGUUU________GUUUGUG .......................................((((.((((((((((((....(((((..(.(((...)))..)..)))))..)))))))))..........))))))) (-19.91 = -19.85 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:04 2006