
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,963,242 – 4,963,406 |
| Length | 164 |
| Max. P | 0.773607 |

| Location | 4,963,242 – 4,963,332 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 72.70 |
| Mean single sequence MFE | -15.50 |
| Consensus MFE | -8.81 |
| Energy contribution | -9.01 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
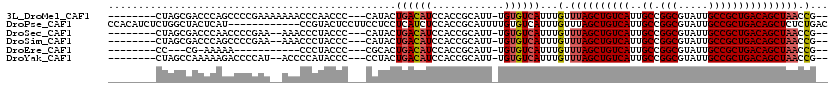
>3L_DroMel_CAF1 4963242 90 + 23771897 UUCUGGAUCUGGAUACACUCAUAUAUCCAUCUCGCUGUGUGUAUGCGGCUCUGUCAGCCUAGCGACCCAGCCCCGAAAAAAACCCAACCC (((.((...((((((........))))))....((((.(.((.(((((((.....))))..)))))))))))).)))............. ( -20.90) >DroSec_CAF1 184022 71 + 1 -----GAUCUGGAUACACUCAUAUAUCCAUCUCGCUGUGU------------GUCAGCCUAGCGACCCAACCCCGAA--AAACCCUACCC -----....((((((........))))))..((((((.((------------....)).))))))............--........... ( -12.90) >DroSim_CAF1 203197 76 + 1 UUCUGGAUCUGGAUACACUCAUAUAUCCAUCUCGCUGUGU------------GUCAGCCUAGCGACCCAGCCCCGAA--AAACCCUACCC ..((((...((((((........))))))..((((((.((------------....)).)))))).)))).......--........... ( -17.70) >DroEre_CAF1 185132 73 + 1 CUCUGGAUCUGGAUACACUCAUAUAUCCAUCUCGCUGUAUG--UGCGGCUCUGUCAGCCC---CG-AAAAA-----------CCCUACCC ..(((((..((((((........))))))..))(((((...--.))))).....)))...---..-.....-----------........ ( -13.90) >DroYak_CAF1 190605 74 + 1 UUCUGGUGCUGGAUACACUCAUAUAUCCAUCUCGCUGU--------------GUCAGCCUAGCCAAAAAGACCCCAU--ACCCCAUACCC ...((((..((((((........))))))....((((.--------------..))))...))))............--........... ( -12.10) >consensus UUCUGGAUCUGGAUACACUCAUAUAUCCAUCUCGCUGUGU____________GUCAGCCUAGCGACCCAGCCCCGAA__AAACCCUACCC .....((..((((((........))))))..))((((.................))))................................ ( -8.81 = -9.01 + 0.20)



| Location | 4,963,300 – 4,963,406 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
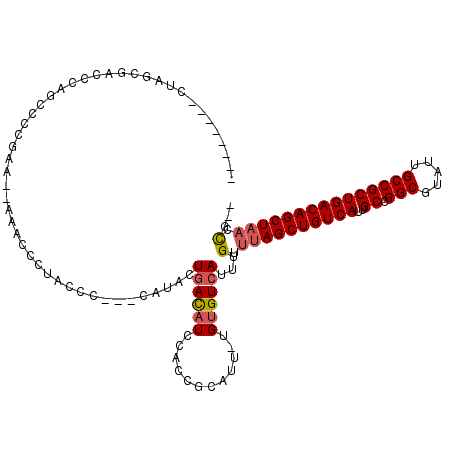
>3L_DroMel_CAF1 4963300 106 + 23771897 --------CUAGCGACCCAGCCCCGAAAAAAACCCAACCC---CAUACUGACAUCCACCGCAUU-UGUGUCAUUUGUUUAGCUGUCAUUGCCGGCGUAUUGCCGCUGACAGCUAACCG-- --------...((......))...................---.....((((((..........-.))))))...(.((((((((((..((.(((.....))))))))))))))).).-- ( -25.10) >DroPse_CAF1 236229 108 + 1 CCACAUCUCUGGCUACUCAU------------CCGUACUCCUUCCUCCUCAUCUCCACCGCAUUUUGUGUCAUUUGUUUAGCUGUCAUUGCCGGCGUAUUGCCGCUGACAGCUCUCUGAC ..........((.(((....------------..)))..)).......(((.......(((.....)))..........((((((((..((.(((.....)))))))))))))...))). ( -19.30) >DroSec_CAF1 184063 104 + 1 --------CUAGCGACCCAACCCCGAA--AAACCCUACCC---CAUACUGACAUCCACCGCAUU-UGUGUCAUUUGUUUAGCUGUCAUUGCCGGCGUAUUGCCGCUGACAGCUAACCG-- --------...................--...........---.....((((((..........-.))))))...(.((((((((((..((.(((.....))))))))))))))).).-- ( -23.90) >DroSim_CAF1 203243 104 + 1 --------CUAGCGACCCAGCCCCGAA--AAACCCUACCC---CAUACUGACAUCCACCGCAUU-UGUGUCAUUUGUUUAGCUGUCAUUGCCGGCGUAUUGCCGCUGACAGCUAACCG-- --------...((......))......--...........---.....((((((..........-.))))))...(.((((((((((..((.(((.....))))))))))))))).).-- ( -25.10) >DroEre_CAF1 185188 91 + 1 --------CC---CG-AAAAA-----------CCCUACCC---CGCACUGACAUCCACCGCAUU-UGUGUCAUUUGUUUAGCUGUCAUUGCCGGCGUAUUGCCGCUGACAGCUAACCG-- --------..---..-.....-----------........---.(((.((((((..........-.))))))..)))((((((((((..((.(((.....)))))))))))))))...-- ( -24.10) >DroYak_CAF1 190649 104 + 1 --------CUAGCCAAAAAGACCCCAU--ACCCCAUACCC---CCUACUGACAUCCACCGCAUU-UGUGUCAUUUGUUUAGCUGUCAUUGCCGGCGUAUUGCCGCUGACAGCUAACCG-- --------...................--...........---.....((((((..........-.))))))...(.((((((((((..((.(((.....))))))))))))))).).-- ( -23.90) >consensus ________CUAGCGACCCAGCCCCGAA__AAACCCUACCC___CAUACUGACAUCCACCGCAUU_UGUGUCAUUUGUUUAGCUGUCAUUGCCGGCGUAUUGCCGCUGACAGCUAACCG__ ................................................((((((............))))))...(.((((((((((..((.(((.....))))))))))))))).)... (-21.26 = -21.65 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:02 2006