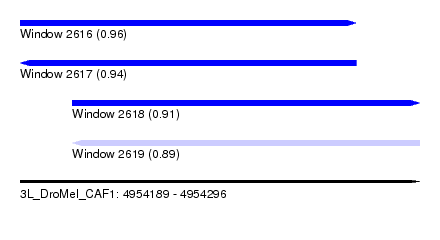
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,954,189 – 4,954,296 |
| Length | 107 |
| Max. P | 0.964146 |

| Location | 4,954,189 – 4,954,279 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -18.61 |
| Energy contribution | -20.33 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
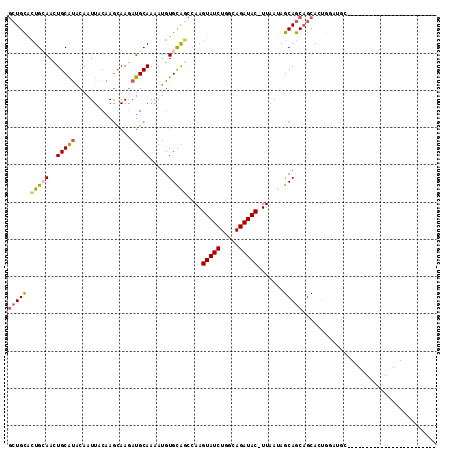
>3L_DroMel_CAF1 4954189 90 + 23771897 ------------------------GCAUCCAGUGCUGCUGCUAUUAA-GUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCUUGCUUGUAAUUGUAUGCAGUUGCAGUGCAGC ------------------------(((...((((.(((.((((...(-(((((.....)))))))))).))))))).)))...(((((((((((((....))))))))).)))). ( -31.20) >DroSec_CAF1 175036 90 + 1 ------------------------GCAGCCAGUGCUGCUGCUAUUAA-GUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCUUGCUUGUAAUUGUAUGCAGUUGCAGUGCAGC ------------------------(((((....)))))......(((-(((((.....))))))))(((((((.....((.....)).((((((((....))))))))))))))) ( -36.10) >DroSim_CAF1 185153 90 + 1 ------------------------GCAGCCAGUGCUGCUGCUAUUAA-GUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCUUGCUUGUAAUUGUAUGCAGUUGCAGUGCAGC ------------------------(((((....)))))......(((-(((((.....))))))))(((((((.....((.....)).((((((((....))))))))))))))) ( -36.10) >DroYak_CAF1 181132 90 + 1 ------------------------GCAUCCAGUGCUGCUGCUAUUAA-GUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCUUGCUUGUAAUUGUAUGCAGUUGCAGUGCAGC ------------------------(((...((((.(((.((((...(-(((((.....)))))))))).))))))).)))...(((((((((((((....))))))))).)))). ( -31.20) >DroMoj_CAF1 247053 80 + 1 --------------------------------UGCAGCUGCUUUUAAUGUAUCUGUCAGAUACUUUGGCACACAUUUUGCAUUUUAUUUGUAAUUGCUCGCAAUU-CACCGCA-- --------------------------------((((((((((......(((((.....)))))...))))......(((((.......)))))..))).)))...-.......-- ( -15.80) >DroAna_CAF1 183465 113 + 1 CCAGCCUCAGCAGCCUCGACUGCCUCUUCCAGUGCUGCUGCUAUUAA-GUAUCUGUCAGAUACUUGGGUGCACAUUUUGCAUCUUGCUUGUAAUUGUAUGCAGUU-CAGUACAGC .......(((((((((.((........)).)).)))))))....(((-(((((.....))))))))((((((.....)))))).((((.(.(((((....)))))-))))).... ( -31.40) >consensus ________________________GCAUCCAGUGCUGCUGCUAUUAA_GUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCUUGCUUGUAAUUGUAUGCAGUUGCAGUGCAGC ...............................(((((((..........(((((.....)))))..(((((((.((.(((((.......)))))..)).))))))))))))))... (-18.61 = -20.33 + 1.72)

| Location | 4,954,189 – 4,954,279 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4954189 90 - 23771897 GCUGCACUGCAACUGCAUACAAUUACAAGCAAGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUAC-UUAAUAGCAGCAGCACUGGAUGC------------------------ (((((.(((((..(((((...............))))).....)))))..(((((((.....)))))-))....))))).(((.....)))------------------------ ( -25.76) >DroSec_CAF1 175036 90 - 1 GCUGCACUGCAACUGCAUACAAUUACAAGCAAGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUAC-UUAAUAGCAGCAGCACUGGCUGC------------------------ (((((((((((.((((............)).)).))))....))))))).(((((((.....)))))-)).......(((((....)))))------------------------ ( -30.70) >DroSim_CAF1 185153 90 - 1 GCUGCACUGCAACUGCAUACAAUUACAAGCAAGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUAC-UUAAUAGCAGCAGCACUGGCUGC------------------------ (((((((((((.((((............)).)).))))....))))))).(((((((.....)))))-)).......(((((....)))))------------------------ ( -30.70) >DroYak_CAF1 181132 90 - 1 GCUGCACUGCAACUGCAUACAAUUACAAGCAAGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUAC-UUAAUAGCAGCAGCACUGGAUGC------------------------ (((((.(((((..(((((...............))))).....)))))..(((((((.....)))))-))....))))).(((.....)))------------------------ ( -25.76) >DroMoj_CAF1 247053 80 - 1 --UGCGGUG-AAUUGCGAGCAAUUACAAAUAAAAUGCAAAAUGUGUGCCAAAGUAUCUGACAGAUACAUUAAAAGCAGCUGCA-------------------------------- --.(((((.-...((((.(((..((((..............))))))))...(((((.....))))).......)))))))).-------------------------------- ( -16.24) >DroAna_CAF1 183465 113 - 1 GCUGUACUG-AACUGCAUACAAUUACAAGCAAGAUGCAAAAUGUGCACCCAAGUAUCUGACAGAUAC-UUAAUAGCAGCAGCACUGGAAGAGGCAGUCGAGGCUGCUGAGGCUGG (((((((..-...(((((...............)))))....))))....(((((((.....)))))-)).....(((((((.((.((........)).))))))))).)))... ( -31.86) >consensus GCUGCACUGCAACUGCAUACAAUUACAAGCAAGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUAC_UUAAUAGCAGCAGCACUGGAUGC________________________ (((((.(((((..(((((...............))))).....)))))....(((((.....))))).......))))).................................... (-17.49 = -17.71 + 0.22)

| Location | 4,954,203 – 4,954,296 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -22.42 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
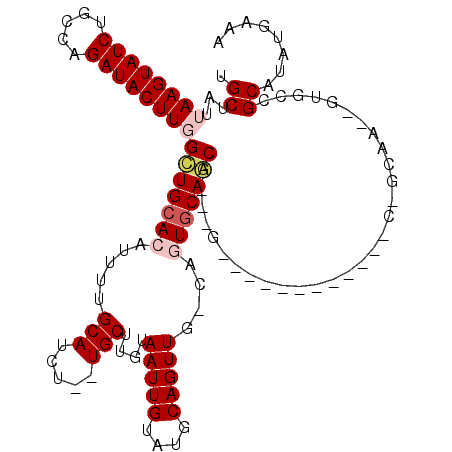
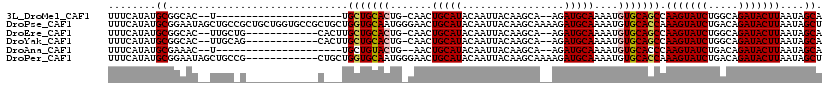
>3L_DroMel_CAF1 4954203 93 + 23771897 UGCUAUUAAGUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCU--UGCUUGUAAUUGUAUGCAGUUG-CAGUGCAGCA---------------------A--GUGCCGCAUAUGAAA (((...((((((((.....))))))))(((((((.....((....--.)).((((((((....))))))-))))))))).---------------------.--.....)))....... ( -32.90) >DroPse_CAF1 225876 119 + 1 AGCUAUUAAGUAUCUGUCAGAUACUUUGGUGCACAUUUUGCAUCUUUUGCUUGUAAUUGUAUGCAGUUCCCAUUGCACCAGCAGCGGCACCAGCAGCGGCAGCUAUUCCGCAUAUGAAA .(((...(((((((.....))))))).((((((((....(((.....))).)))..((((.((((((....))))))...))))..)))))))).((((........))))........ ( -33.60) >DroEre_CAF1 176104 102 + 1 UGCUAUUAAGUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCU--UGCUUGUAAUUGUAUGCAGUUG-CAGUGCAGCAAGUG------------CAGCAA--GUGCCGCAUAUGAAA (((...((((((((.....))))))))(((((((...((((....--((((((((((((....))))))-))).))))))))))------------))))..--.....)))....... ( -37.30) >DroYak_CAF1 181146 102 + 1 UGCUAUUAAGUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCU--UGCUUGUAAUUGUAUGCAGUUG-CAGUGCAGCAAGUG------------CUGCAA--GUGCCGCAUAUGAAA ......((((((((.....))))))))(((((((.....((....--.)).((((((((....))))))-)))))))))..(((------------(.((..--..)).))))...... ( -37.00) >DroAna_CAF1 183503 92 + 1 UGCUAUUAAGUAUCUGUCAGAUACUUGGGUGCACAUUUUGCAUCU--UGCUUGUAAUUGUAUGCAGUU--CAGUACAGCA---------------------A--GUUUCGCAUAUGAAA (((..(((((((((.....)))))))))..)))(((..(((..((--((((.(((((((....)))).--...)))))))---------------------)--)....))).)))... ( -25.10) >DroPer_CAF1 210051 107 + 1 AGCUAUUAAGUAUCUGUCAGAUACUUUGGUGCACAUUUUGCAUCUUUUGCUUGUAAUUGUAUGCAGUUCCCAUUGCACCAGCAG------------CGGCAGCUAUUCCGCAUAUGAAA .(((...(((((((.....))))))).((((((....(((((((..(((....)))..).)))))).......))))))))).(------------(((........))))........ ( -30.70) >consensus UGCUAUUAAGUAUCUGCCAGAUACUUGGCUGCACAUUUUGCAUCU__UGCUUGUAAUUGUAUGCAGUUG_CAGUGCAGCA___G____________C_GCAA__GUGCCGCAUAUGAAA .((...((((((((.....))))))))(((((((.....(((.....)))....(((((....)))))....)))))))..............................))........ (-22.42 = -23.08 + 0.67)


| Location | 4,954,203 – 4,954,296 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -21.05 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
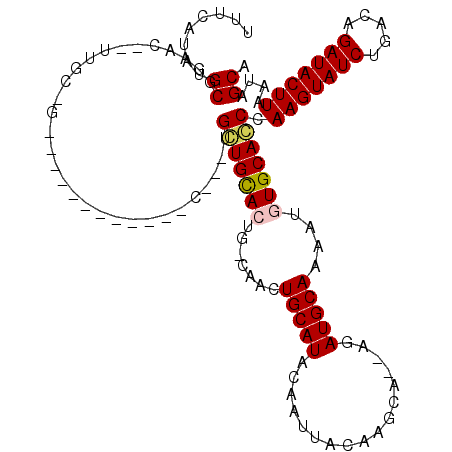
>3L_DroMel_CAF1 4954203 93 - 23771897 UUUCAUAUGCGGCAC--U---------------------UGCUGCACUG-CAACUGCAUACAAUUACAAGCA--AGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUACUUAAUAGCA .......(((.....--.---------------------.(((((((((-((.((((............)).--)).))))....))))))).(((((((.....)))))))....))) ( -26.30) >DroPse_CAF1 225876 119 - 1 UUUCAUAUGCGGAAUAGCUGCCGCUGCUGGUGCCGCUGCUGGUGCAAUGGGAACUGCAUACAAUUACAAGCAAAAGAUGCAAAAUGUGCACCAAAGUAUCUGACAGAUACUUAAUAGCU ........((((..((((.......))))...)))).(((((((((..(....)(((((.................))))).....)))))).(((((((.....)))))))...))). ( -34.03) >DroEre_CAF1 176104 102 - 1 UUUCAUAUGCGGCAC--UUGCUG------------CACUUGCUGCACUG-CAACUGCAUACAAUUACAAGCA--AGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUACUUAAUAGCA .......((((((..--..))))------------))...(((((((((-((.((((............)).--)).))))....))))))).(((((((.....)))))))....... ( -32.60) >DroYak_CAF1 181146 102 - 1 UUUCAUAUGCGGCAC--UUGCAG------------CACUUGCUGCACUG-CAACUGCAUACAAUUACAAGCA--AGAUGCAAAAUGUGCAGCCAAGUAUCUGGCAGAUACUUAAUAGCA .......((((((..--.(((((------------(....)))))).((-((..(((((.............--..))))).....)))))))(((((((.....)))))))....))) ( -31.06) >DroAna_CAF1 183503 92 - 1 UUUCAUAUGCGAAAC--U---------------------UGCUGUACUG--AACUGCAUACAAUUACAAGCA--AGAUGCAAAAUGUGCACCCAAGUAUCUGACAGAUACUUAAUAGCA .......(((....(--(---------------------(((((((...--.............))).))))--)).((((.....))))...(((((((.....)))))))....))) ( -18.59) >DroPer_CAF1 210051 107 - 1 UUUCAUAUGCGGAAUAGCUGCCG------------CUGCUGGUGCAAUGGGAACUGCAUACAAUUACAAGCAAAAGAUGCAAAAUGUGCACCAAAGUAUCUGACAGAUACUUAAUAGCU ........((((........)))------------).(((((((((..(....)(((((.................))))).....)))))).(((((((.....)))))))...))). ( -27.93) >consensus UUUCAUAUGCGGAAC__UUGC_G____________C___UGCUGCACUG_CAACUGCAUACAAUUACAAGCA__AGAUGCAAAAUGUGCACCCAAGUAUCUGACAGAUACUUAAUAGCA ........((..............................(((((((.......(((((.................)))))....))))))).(((((((.....)))))))....)). (-21.05 = -21.08 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:56 2006