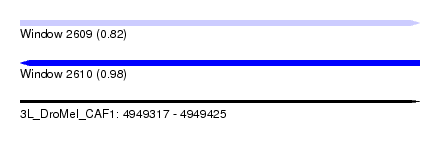
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,949,317 – 4,949,425 |
| Length | 108 |
| Max. P | 0.984483 |

| Location | 4,949,317 – 4,949,425 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -17.78 |
| Energy contribution | -18.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
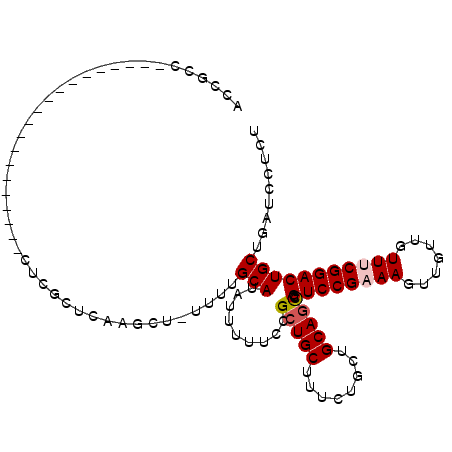
>3L_DroMel_CAF1 4949317 108 + 23771897 ACCGCCCACCAACCCGCCAACAUUUUCUCGCUCAAGCU-UUUUGCAUAUUUUUCCGCUGCUUUCUGCUGCAACCGUCCGAAAGUUGUUGUUGCGGACUGCUGAUCCUCU ......((.((..((((((((((((((..((....((.-....))..........((.((.....)).))....))..))))).)))))..))))..)).))....... ( -22.60) >DroPse_CAF1 218947 84 + 1 GUCUCC--------------------GUUUAUCAUUC--UUUUGCAUAUUUUUCCGCUGCUUUCCGCUGCAGUCGUCCGAAAGAUG---UUUCGGACUGCUGAUCCUAU ......--------------------...........--....(((.........(((((........))))).((((((((....---)))))))))))......... ( -19.30) >DroSec_CAF1 170308 88 + 1 ACCGCC--------------------CUCGCUCAAGCU-UUUUGCAUAUUUUUCCGCUGCUUUCUGCUGCAGCCGUCCGAAAGUUGUUGUUUCGGACUGCUGAUCCUCU ......--------------------...((....)).-....(((.........(((((........))))).((((((((.......)))))))))))......... ( -22.10) >DroSim_CAF1 180350 88 + 1 ACCGCC--------------------CUCGCUCAAGCU-UUUUGCAUAUUUUUCCGCUGCUUUCUGCUGCAGCCGUCCGAAAGUUGUUGUUUCGGACUGCUGAUCCUCU ......--------------------...((....)).-....(((.........(((((........))))).((((((((.......)))))))))))......... ( -22.10) >DroYak_CAF1 176147 108 + 1 UCAGCCCGCCAA-ACGCCCACGUUUUCUCGCUCAAGCCUUUUUGCAUAUUUUUCCGCUGCUUUCUGCUGCAGCCGUCCGAAAGUUGUUGUUGCGGACUGCUGAUCCUCU (((((((((.((-(((....)))))..(((..(..((......))..........(((((........))))).)..)))...........))))...)))))...... ( -25.60) >consensus ACCGCC____________________CUCGCUCAAGCU_UUUUGCAUAUUUUUCCGCUGCUUUCUGCUGCAGCCGUCCGAAAGUUGUUGUUUCGGACUGCUGAUCCUCU ...........................................(((.........(((((........))))).((((((((.......)))))))))))......... (-17.78 = -18.22 + 0.44)

| Location | 4,949,317 – 4,949,425 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
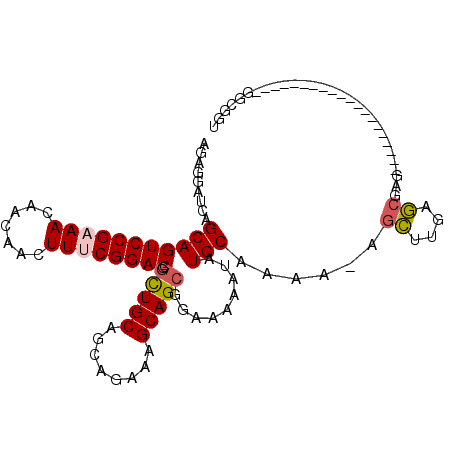
>3L_DroMel_CAF1 4949317 108 - 23771897 AGAGGAUCAGCAGUCCGCAACAACAACUUUCGGACGGUUGCAGCAGAAAGCAGCGGAAAAAUAUGCAAAA-AGCUUGAGCGAGAAAAUGUUGGCGGGUUGGUGGGCGGU ....(..((.((..((((..(((((..(((((..(.(((((........))))).)........((....-.)).....)))))...)))))))))..)).))..)... ( -31.40) >DroPse_CAF1 218947 84 - 1 AUAGGAUCAGCAGUCCGAAA---CAUCUUUCGGACGACUGCAGCGGAAAGCAGCGGAAAAAUAUGCAAAA--GAAUGAUAAAC--------------------GGAGAC .........(((((((((((---....))))))))..((((.((.....)).)))).......)))....--...........--------------------(....) ( -21.90) >DroSec_CAF1 170308 88 - 1 AGAGGAUCAGCAGUCCGAAACAACAACUUUCGGACGGCUGCAGCAGAAAGCAGCGGAAAAAUAUGCAAAA-AGCUUGAGCGAG--------------------GGCGGU .....(((.(((((((((((.......)))))))).(((((........))))).........)))....-.((((......)--------------------)))))) ( -26.20) >DroSim_CAF1 180350 88 - 1 AGAGGAUCAGCAGUCCGAAACAACAACUUUCGGACGGCUGCAGCAGAAAGCAGCGGAAAAAUAUGCAAAA-AGCUUGAGCGAG--------------------GGCGGU .....(((.(((((((((((.......)))))))).(((((........))))).........)))....-.((((......)--------------------)))))) ( -26.20) >DroYak_CAF1 176147 108 - 1 AGAGGAUCAGCAGUCCGCAACAACAACUUUCGGACGGCUGCAGCAGAAAGCAGCGGAAAAAUAUGCAAAAAGGCUUGAGCGAGAAAACGUGGGCGU-UUGGCGGGCUGA ..........((((((((.........(((((..(.(((((........))))).)........((......)).....)))))(((((....)))-)).)))))))). ( -33.70) >consensus AGAGGAUCAGCAGUCCGAAACAACAACUUUCGGACGGCUGCAGCAGAAAGCAGCGGAAAAAUAUGCAAAA_AGCUUGAGCGAG____________________GGCGGU .........(((((((((((.......)))))))).(((((........))))).........)))......((....))............................. (-19.34 = -19.66 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:47 2006