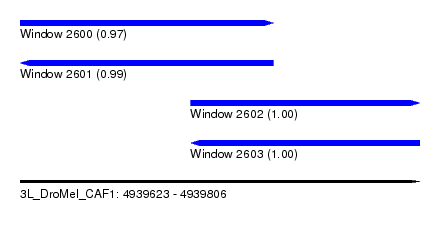
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,939,623 – 4,939,806 |
| Length | 183 |
| Max. P | 0.999381 |

| Location | 4,939,623 – 4,939,739 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -15.97 |
| Energy contribution | -15.58 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4939623 116 + 23771897 AUGGCCAGAAGGGAA--GGGGGUAUGGGAAAAUGGAUGAGAGAGAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCAG (((.((.........--..)).))).......((..((((.(((.((.......))........((((((....)))))).((((((((((....)))))))))).))))--)))..)). ( -27.80) >DroSim_CAF1 170811 117 + 1 UUGGCCAGAAGGGAA--GG-GGUAUGGGAAAAUGGCUGAAAGAGAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAG (..((((........--..-............))))..)..(((((.(((...(((((((((....)).)))))))......(((((((((....))))))))))))..)))))...... ( -29.55) >DroEre_CAF1 162114 118 + 1 AUGGCCAGAAGGGAAAAGGGGGAGUGGGAAAAUGGCUGAAAGAGAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCAG ....((....)).........((((((((.....(((.......)))(((...(((((((((....)).)))))))......(((((((((....)))))))))))))))--.))))).. ( -29.90) >DroYak_CAF1 166193 109 + 1 AUGGCCAGAAGAGUAG---------GGGAAAAUGGCUGAAAGAGAGCGUUACAAACAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCAG ..........((((.(---------((((.....(((.......)))(((...(((((((((....)).)))))))......(((((((((....)))))))))))))))--)))))).. ( -29.70) >DroMoj_CAF1 225579 97 + 1 UUUACAAGGAG--UU----------GGG------CGUGACUUAGGGCGUGUC-AGCAAAUGAAAAAUCAAUUUGUUGUUUAUGCAUAAAUCAUGUGAUUUAUGUGACUUC--UAA--CAG ..........(--((----------(((------.((.((.....(((((.(-(((((((((....)).))))))))..)))))(((((((....))))))))).)).))--)))--).. ( -26.80) >DroAna_CAF1 169786 100 + 1 AUUGCGAGGAU--------------GAGAGAAG----CAGUGUGAGCAUUACAAGAAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCGG ....((((((.--------------(((.....----.((((....)))).(((.(((.(((....))).))).)))....((((((((((....)))))))))).))).--)).)))). ( -26.90) >consensus AUGGCCAGAAGGGAA__________GGGAAAAUGGCUGAAAGAGAGCGUUACAAGCAAAUGAAAAAUCAAUUUGUUGAUUAUGCAUAAAUCAUAAGAUUUAUGCGACUCC__UCACUCAG ...........................................((((......(((((((((....)).)))))))......(((((((((....)))))))))).)))........... (-15.97 = -15.58 + -0.39)
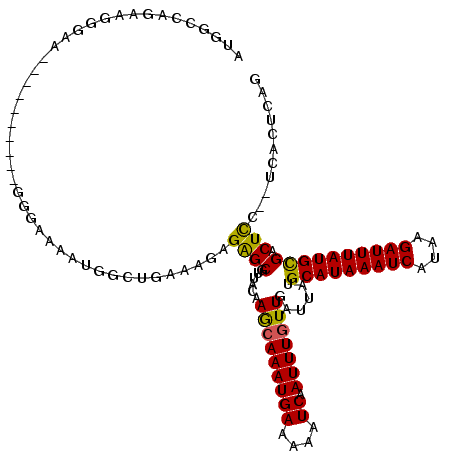
| Location | 4,939,623 – 4,939,739 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -16.46 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4939623 116 - 23771897 CUGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUCUCUCUCAUCCAUUUUCCCAUACCCCC--UUCCCUUCUGGCCAU .((((.((--(.(((.(((((((((....))))))))).....((((((((.((....))))))).)))....)))))).))))...................--...((....)).... ( -25.40) >DroSim_CAF1 170811 117 - 1 CUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUCUCUUUCAGCCAUUUUCCCAUACC-CC--UUCCCUUCUGGCCAA .....((((((((((.(((((((((....))))))))).....((((((((.((....))))))).)))....)))).))))))((((............-..--........))))... ( -28.45) >DroEre_CAF1 162114 118 - 1 CUGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUCUCUUUCAGCCAUUUUCCCACUCCCCCUUUUCCCUUCUGGCCAU ..(((((.--((((..(((((((((....))))))))).....((((((((.((....))))))).)))....(((.......)))....))))))))).........((....)).... ( -26.80) >DroYak_CAF1 166193 109 - 1 CUGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGUUUGUAACGCUCUCUUUCAGCCAUUUUCCC---------CUACUCUUCUGGCCAU .((((.((--(.(((.(((((((((....)))))))))...((.(((((((.((....))))))))).))...)))))).))))((((.......---------.........))))... ( -26.59) >DroMoj_CAF1 225579 97 - 1 CUG--UUA--GAAGUCACAUAAAUCACAUGAUUUAUGCAUAAACAACAAAUUGAUUUUUCAUUUGCU-GACACGCCCUAAGUCACG------CCC----------AA--CUCCUUGUAAA .((--(((--(......((((((((....)))))))).........(((((.((....)))))))))-))))..............------...----------..--........... ( -14.50) >DroAna_CAF1 169786 100 - 1 CCGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUUCUUGUAAUGCUCACACUG----CUUCUCUC--------------AUCCUCGCAAU ....((((--(((...(((((((((....))))))))).....(((.(((.(((....))).))).)))....((.......)----).......--------------.)))))))... ( -26.50) >consensus CUGAGUGA__GGAGUCGCAUAAAUCUUAUGAUUUAUGCAUAAUCAACAAAUUGAUUUUUCAUUUGCUUGUAACGCUCUCUUUCAGCCAUUUUCCC__________UUCCCUUCUGGCCAU ..(((((.........(((((((((....))))))))).....((((((((.((....))))))).)))...)))))........................................... (-16.46 = -17.35 + 0.89)

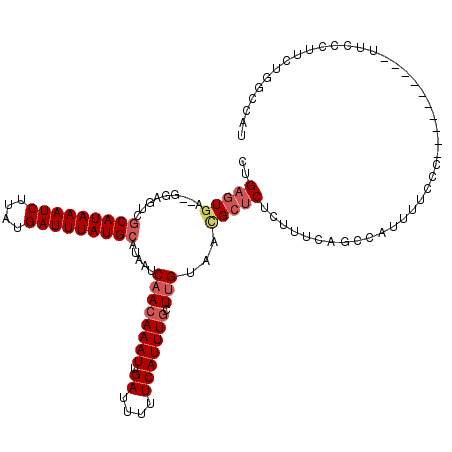

| Location | 4,939,701 – 4,939,806 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4939701 105 + 23771897 AUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUUCAUGUCAGUAUCACAAAAAC ..(((((((((....)))))))))(((((.--...............)))))...........(((.((....)))))(((((.(.....).))))).......... ( -19.79) >DroSec_CAF1 160842 107 + 1 AUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUCCAUGUCAGAAUCACAAAAAC ..(((((((((....)))))))))(((.......(((((.......)))))............(((((.............)).)))...))).............. ( -18.92) >DroSim_CAF1 170888 107 + 1 AUGCAUAAAUCAUAAGAUUUAUGCGACUCCUCUCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUUCAUGUCAGAAUCACAAAAAC ..(((((((((....)))))))))(((.......(((((.......)))))............(((((.............)))..))..))).............. ( -18.22) >DroEre_CAF1 162194 105 + 1 AUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCAGCAUUUAUGAGUCAUUUAUCAAGCGAGCAAACUUUUCUCUACUGCCUUCAUGUCAGAAUCACAAAAAC ..(((((((((....)))))))))(((...--..(((((.......)))))............(((((.............)))..))..))).............. ( -18.22) >DroYak_CAF1 166264 105 + 1 AUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCAGCAUUUAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCCACUGCCUUCAUGUCAGAAUCACAAAAAC ..(((((((((....)))))))))(((...--..(((((.......)))))............(((((.............)))..))..))).............. ( -18.22) >DroAna_CAF1 169848 103 + 1 AUGCAUAAAUCAUAAGAUUUAUGCGACUCC--UCACUCGGCAUUCAUGAGUCAUUUAUCAAACGGGCCAGCUC--CUUUUUUGCCUUCGUGUCAAAAUCACAAAAUC .((((((((((....)))))))))).....--......((((....(((........)))...(((....)))--......))))...(((.......)))...... ( -21.10) >consensus AUGCAUAAAUCAUAAGAUUUAUGCGACUCC__UCACUCAGCAUUCAUGAGUCAUUUAUCAAACGAGCAAACUUUUCUCUACUGCCUUCAUGUCAGAAUCACAAAAAC ..(((((((((....)))))))))(((.......(((((.......)))))............(((.........)))............))).............. (-17.65 = -17.23 + -0.41)

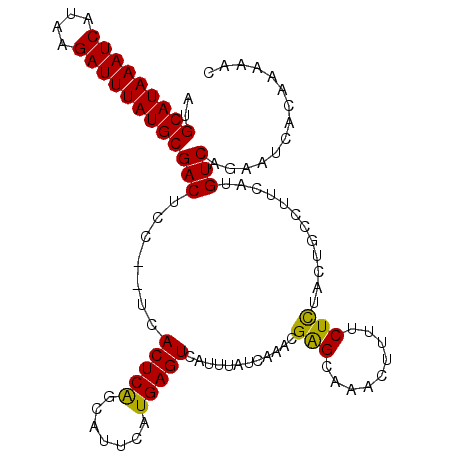
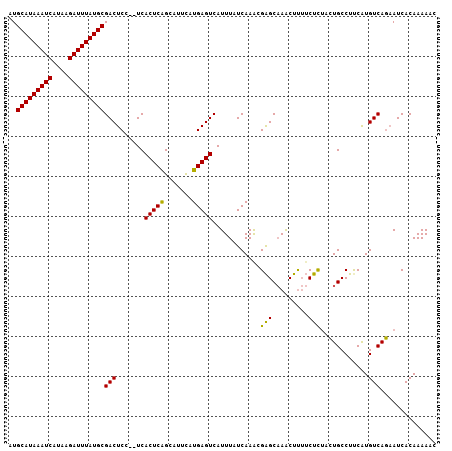
| Location | 4,939,701 – 4,939,806 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -26.05 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4939701 105 - 23771897 GUUUUUGUGAUACUGACAUGAAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAU (((((..((.......))..)))))((((((......)))))).....(((...(.(((((.......))))).)--...)))(((((((((....))))))))).. ( -26.60) >DroSec_CAF1 160842 107 - 1 GUUUUUGUGAUUCUGACAUGGAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAU ........((((((..(((...((((...(((....(((((.((....)))))))..))).....))))..)))...))))))(((((((((....))))))))).. ( -28.60) >DroSim_CAF1 170888 107 - 1 GUUUUUGUGAUUCUGACAUGAAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGAGAGGAGUCGCAUAAAUCUUAUGAUUUAUGCAU ........((((((..(((...((((...(((....(((((.((....)))))))..))).....))))..)))...))))))(((((((((....))))))))).. ( -28.60) >DroEre_CAF1 162194 105 - 1 GUUUUUGUGAUUCUGACAUGAAGGCAGUAGAGAAAAGUUUGCUCGCUUGAUAAAUGACUCAUAAAUGCUGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAU ........((((((..(((..((((((((((......)))))).)))).....)))(((((.......)))))..--))))))(((((((((....))))))))).. ( -30.50) >DroYak_CAF1 166264 105 - 1 GUUUUUGUGAUUCUGACAUGAAGGCAGUGGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUAAAUGCUGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAU ........((((((..(((...((((...(((....(((((.((....)))))))..))).....))))..))).--))))))(((((((((....))))))))).. ( -29.80) >DroAna_CAF1 169848 103 - 1 GAUUUUGUGAUUUUGACACGAAGGCAAAAAAG--GAGCUGGCCCGUUUGAUAAAUGACUCAUGAAUGCCGAGUGA--GGAGUCGCAUAAAUCUUAUGAUUUAUGCAU ........((((((..(((...((((.....(--(.(....)))(((........))).......))))..))).--))))))(((((((((....))))))))).. ( -28.40) >consensus GUUUUUGUGAUUCUGACAUGAAGGCAGUAGAGAAAAGUUUGCUCGUUUGAUAAAUGACUCAUGAAUGCUGAGUGA__GGAGUCGCAUAAAUCUUAUGAUUUAUGCAU ........((((((..(((...((((...(((....((((((......).)))))..))).....))))..)))...))))))(((((((((....))))))))).. (-26.05 = -26.30 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:40 2006