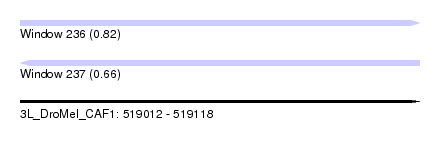
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 519,012 – 519,118 |
| Length | 106 |
| Max. P | 0.821666 |

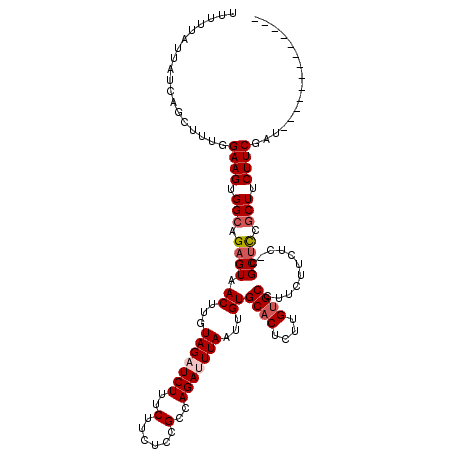
| Location | 519,012 – 519,118 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -20.61 |
| Energy contribution | -20.89 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 519012 106 + 23771897 -------------AUCGAAGAAGCGGAGCG-GAGAAGAAGGCACAAGAGUGCACAAUUAAAUCUGGCGGAGAAGAAAGAUCUACAAGUUACUCUGCCACUUCCAAAGCUGAUAAUAAAAA -------------.(((......)))(((.-(.((((...((((....))))...........((((((((...................)))))))))))))...)))........... ( -24.01) >DroSec_CAF1 98784 106 + 1 -------------AUCGAAGAAGCGAAGCG-GAGAAGACGGCACAAGAGUGCACAAUUAAAUCUGGCGGAGAAGAAAGAUCUACAAGUUACUCUGCCACUUCCAAAGCUGAUAAUAAAAA -------------.(((......)))(((.-(.((((...((((....))))...........((((((((...................)))))))))))))...)))........... ( -24.81) >DroSim_CAF1 102734 106 + 1 -------------AUCGAAGAAGCGGAGCU-GAGAAGACGGCACAAGAGUGCACAAUUAAAUCUGGCGGAGAAGAAAGAUCUACAAGUUACUCUGCCACUUCCAAAGCUGAUAAUAAAAA -------------.(((......)))((((-..((((...((((....))))...........((((((((...................))))))))))))...))))........... ( -25.21) >DroEre_CAF1 102633 112 + 1 GGAG--------AAUCGAAGAAGCGGAGCGAGAGAAGAAGGCCCAAGAGUGCACAAUUAAUUCUGGCGGAGAAGAAAGAUCUACAAGUUACUCUGCCACUUCCAAAGCUGAUAAUAAAAA ....--------..(((......)))(((....((((........(((((.........)))))(((((((...................))))))).))))....)))........... ( -20.01) >DroYak_CAF1 104819 120 + 1 GGAGAAUCAGAGAAUCGAAGAAGUGGAGCGAGCGAAGAAGGCACAAGAGUGCACAAUUAAUUCUGGCGGAGAAGAAAGAUCUACAAGUUACACAGCCACUUCCAAAGCUGAUAAUAAAAA .....(((((.........(((((((....(((..(((..((((....)))).........(((..(......)..))))))....)))......))))))).....)))))........ ( -23.64) >consensus _____________AUCGAAGAAGCGGAGCG_GAGAAGAAGGCACAAGAGUGCACAAUUAAAUCUGGCGGAGAAGAAAGAUCUACAAGUUACUCUGCCACUUCCAAAGCUGAUAAUAAAAA ..............(((......)))(((....((((...((((....))))...........((((((((...................))))))))))))....)))........... (-20.61 = -20.89 + 0.28)

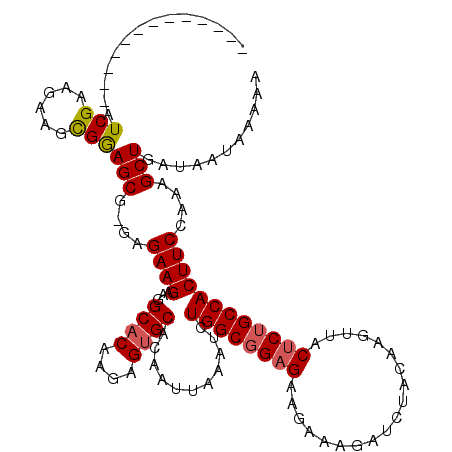
| Location | 519,012 – 519,118 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 519012 106 - 23771897 UUUUUAUUAUCAGCUUUGGAAGUGGCAGAGUAACUUGUAGAUCUUUCUUCUCCGCCAGAUUUAAUUGUGCACUCUUGUGCCUUCUUCUC-CGCUCCGCUUCUUCGAU------------- ..................((((.(((.((((.((...(((((((..(......)..)))))))...))((((....)))).........-.)))).))).))))...------------- ( -23.60) >DroSec_CAF1 98784 106 - 1 UUUUUAUUAUCAGCUUUGGAAGUGGCAGAGUAACUUGUAGAUCUUUCUUCUCCGCCAGAUUUAAUUGUGCACUCUUGUGCCGUCUUCUC-CGCUUCGCUUCUUCGAU------------- ...........(((...((((((((((((((.((...(((((((..(......)..)))))))...))..))))...))))).))))).-.)))(((......))).------------- ( -24.10) >DroSim_CAF1 102734 106 - 1 UUUUUAUUAUCAGCUUUGGAAGUGGCAGAGUAACUUGUAGAUCUUUCUUCUCCGCCAGAUUUAAUUGUGCACUCUUGUGCCGUCUUCUC-AGCUCCGCUUCUUCGAU------------- ...........((((..((((((((((((((.((...(((((((..(......)..)))))))...))..))))...))))).))))).-)))).............------------- ( -25.30) >DroEre_CAF1 102633 112 - 1 UUUUUAUUAUCAGCUUUGGAAGUGGCAGAGUAACUUGUAGAUCUUUCUUCUCCGCCAGAAUUAAUUGUGCACUCUUGGGCCUUCUUCUCUCGCUCCGCUUCUUCGAUU--------CUCC ...........(((...(((((.((((((((.....(.(((.......))).)(((((......))).)))))))...)))..)))))...)))..............--------.... ( -19.00) >DroYak_CAF1 104819 120 - 1 UUUUUAUUAUCAGCUUUGGAAGUGGCUGUGUAACUUGUAGAUCUUUCUUCUCCGCCAGAAUUAAUUGUGCACUCUUGUGCCUUCUUCGCUCGCUCCACUUCUUCGAUUCUCUGAUUCUCC ........(((((....((((((((..(((......(.(((.......))).)((.((((......(.((((....)))))))))..)).))).))))))))........)))))..... ( -23.00) >consensus UUUUUAUUAUCAGCUUUGGAAGUGGCAGAGUAACUUGUAGAUCUUUCUUCUCCGCCAGAUUUAAUUGUGCACUCUUGUGCCUUCUUCUC_CGCUCCGCUUCUUCGAU_____________ ..................((((.(((.((((.((...(((((((..(......)..)))))))...))((((....))))...........)))).))).))))................ (-18.86 = -19.70 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:15 2006