| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,824,001 – 4,824,101 |
| Length | 100 |
| Max. P | 0.930363 |

| Location | 4,824,001 – 4,824,101 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
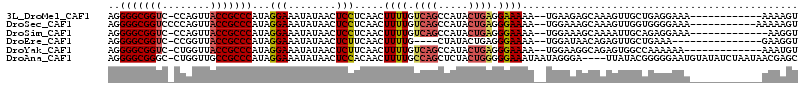
>3L_DroMel_CAF1 4824001 100 + 23771897 AGGGGCGGUC-CCAGUUACCGCCCAUAGGAAAUAUAACUCCUCAACUUUUGUCAGCCAUACUGAGGAAAAA--UGAAGAGCAAAGUUGCUGAGGAAA------------AAAAGU ..(((((((.-......)))))))..............(((((...((((.((((.....)))).))))..--.....((((....)))))))))..------------...... ( -30.30) >DroSec_CAF1 51574 102 + 1 AGGGGCGGUCCCCAGUUACCGCCCAUAGGAAAUAUAACUCCUCAACUUUUGUCAGCCAUACUGAGGGAAAA--UGGAAAGCAAAGUUGGUGGGGAAA-----------AAAAAGU ..(((((((........))))))).............((((((((((((.((...((((...........)--)))...)))))))))).))))...-----------....... ( -31.00) >DroSim_CAF1 54025 99 + 1 AGGGGCGGUC-CCAGUUACCGCCCAUAGGAAAUAUAACUCCUCAACUUUUGUCAGCCAUACUGAGGGAAAA--UGGAAAGCAAAAUUGCAGAGGAAA-------------AAGGU ..(((((((.-......)))))))..............(((((...((((.((((.....)))).))))..--......(((....))).)))))..-------------..... ( -29.30) >DroEre_CAF1 52468 93 + 1 AGGGGCGGUC-CCGGUUACCGCCCAUAGGAAAUAUAACUCUUCAACUUUUG----CUAUACUGAGGGAAAA--UGGAUAACAGAGUUGCUGAAA---------------GAAGGU ..(((((((.-......)))))))..........(((((((((...((((.----((......)).)))).--..))....)))))))......---------------...... ( -22.30) >DroYak_CAF1 52450 99 + 1 AGGGGCGGUC-CUGGUUACCGCCCAUAGGAAAUAUAACUCUUCAACUUUUGUCAGCCAUACUGAGGGAAAA--UGGAAGGCAGAGUGGCCAAAAAA-------------AAAUGU ..(((((((.-......)))))))................((((..((((.((((.....)))).))))..--)))).(((......)))......-------------...... ( -26.20) >DroAna_CAF1 56139 110 + 1 AGGGGCGGGC-CUGGUUGCCGCCCAUAGGAAAUAUAACUCCACAACUUUUGCCAGCUCUACUGGGGGAAAUAAUAGGGA----UUAUACGGGGGAAUGUAUAUCUAAUAACGAGC ..((((((..-.......))))))...(((.(((((.((((.....((((.((((.....)))).))))(((((....)----))))..))))...))))).))).......... ( -29.50) >consensus AGGGGCGGUC_CCAGUUACCGCCCAUAGGAAAUAUAACUCCUCAACUUUUGUCAGCCAUACUGAGGGAAAA__UGGAAAGCAAAGUUGCUGAGGAAA____________AAAAGU ..(((((((........)))))))...(((........))).....((((.((((.....)))).)))).............................................. (-18.85 = -19.10 + 0.25)
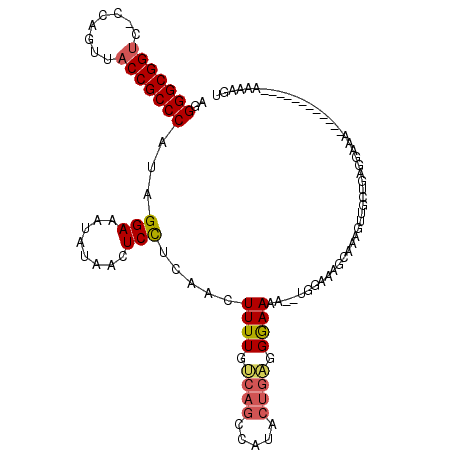

| Location | 4,824,001 – 4,824,101 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -17.56 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
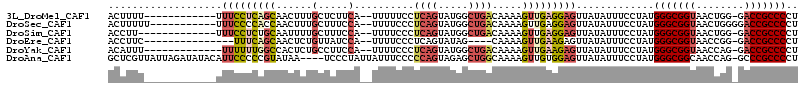
>3L_DroMel_CAF1 4824001 100 - 23771897 ACUUUU------------UUUCCUCAGCAACUUUGCUCUUCA--UUUUUCCUCAGUAUGGCUGACAAAAGUUGAGGAGUUAUAUUUCCUAUGGGCGGUAACUGG-GACCGCCCCU ......------------................((((((((--.((((..((((.....))))..)))).))))))))............(((((((......-.))))))).. ( -29.30) >DroSec_CAF1 51574 102 - 1 ACUUUUU-----------UUUCCCCACCAACUUUGCUUUCCA--UUUUCCCUCAGUAUGGCUGACAAAAGUUGAGGAGUUAUAUUUCCUAUGGGCGGUAACUGGGGACCGCCCCU .......-----------.........(((((((.....(((--(...........))))......)))))))(((((......)))))..(((((((........))))))).. ( -26.90) >DroSim_CAF1 54025 99 - 1 ACCUU-------------UUUCCUCUGCAAUUUUGCUUUCCA--UUUUCCCUCAGUAUGGCUGACAAAAGUUGAGGAGUUAUAUUUCCUAUGGGCGGUAACUGG-GACCGCCCCU .....-------------.((((((.((..((((((...(((--(...........))))..).))))))).)))))).............(((((((......-.))))))).. ( -26.10) >DroEre_CAF1 52468 93 - 1 ACCUUC---------------UUUCAGCAACUCUGUUAUCCA--UUUUCCCUCAGUAUAG----CAAAAGUUGAAGAGUUAUAUUUCCUAUGGGCGGUAACCGG-GACCGCCCCU ......---------------.(((..(((((.((((((.(.--..........).))))----))..)))))..))).............(((((((......-.))))))).. ( -21.20) >DroYak_CAF1 52450 99 - 1 ACAUUU-------------UUUUUUGGCCACUCUGCCUUCCA--UUUUCCCUCAGUAUGGCUGACAAAAGUUGAAGAGUUAUAUUUCCUAUGGGCGGUAACCAG-GACCGCCCCU .((.((-------------(((.(..((((..(((.......--........)))..))))..).))))).))..................(((((((......-.))))))).. ( -26.16) >DroAna_CAF1 56139 110 - 1 GCUCGUUAUUAGAUAUACAUUCCCCCGUAUAA----UCCCUAUUAUUUCCCCCAGUAGAGCUGGCAAAAGUUGUGGAGUUAUAUUUCCUAUGGGCGGCAACCAG-GCCCGCCCCU ..........................((((((----..((.((.((((...((((.....))))...)))).))))..)))))).......(((((((......-).)))))).. ( -24.20) >consensus ACUUUU____________UUUCCUCAGCAACUUUGCUUUCCA__UUUUCCCUCAGUAUGGCUGACAAAAGUUGAGGAGUUAUAUUUCCUAUGGGCGGUAACCGG_GACCGCCCCU ...................(((((((((......(.....)..........((((.....)))).....))))))))).............(((((((........))))))).. (-17.56 = -18.48 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:13 2006