| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,806,335 – 4,806,446 |
| Length | 111 |
| Max. P | 0.967264 |

| Location | 4,806,335 – 4,806,446 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.09 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.39 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.61 |
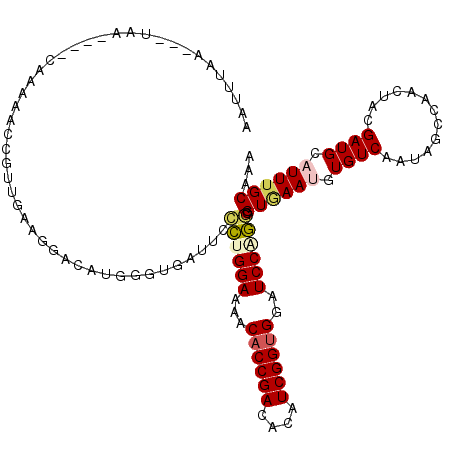
| Structure conservation index | 0.55 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
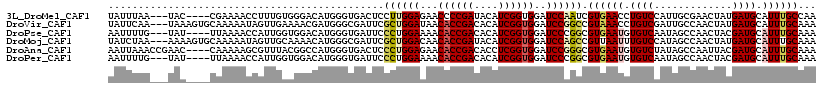
>3L_DroMel_CAF1 4806335 111 - 23771897 UAUUUAA---UAC----CGAAAACCUUUGUGGGACAUGGGUGACUCCUUGGAGAACCCCGAUACAUCGGUGGAUCCAAUCGUGAACCUGUCCAUUGCGAACUAUGAUGCAUUUGCCAA .......---...----........(((((.(((((.((.(.((...(((((...((((((....)))).)).)))))..)).).)))))))...))))).......((....))... ( -26.40) >DroVir_CAF1 46779 115 - 1 UAUUCAA---UAAAGUGCAAAAAUAGUUGAAAACGAUGGGCGAUUCGCUGGAUAACACCGACACAUCGGUGGAUCCGGCCGUAAACCUGUCGAUUGCCAACUAUGAUGCAUUUGCAAA .......---..(((((((...(((((((..(((((((((......(((((((..((((((....)))))).)))))))......))))))).))..)))))))..)))))))..... ( -43.30) >DroPse_CAF1 69175 111 - 1 AAUUUUG---UAU----UUAAAACCAUUGGUGGACAUGGGUGAUUCCCUGGAAAACACCGACACAUCGGUGGAUCCCGGCGUGAAUGUGUCAAUAGCCAACUACGAUGCAUUUGCAAA .......---...----......(((((((((....(.((((.(((....)))..)))).)..)))))))))........(..((((((((..(((....))).))))))))..)... ( -30.50) >DroMoj_CAF1 67360 115 - 1 UAUCUAA---AAAAGUGCAAAAAUAGUUGCAAAACAUGGGCGAUUCGCUGGACAACACCGAUACAUCGGUGGAUCCAGCCGUUAAUUUGUCCAUAGCCAACUAUGAUGCAUUUGCAAA .......---..(((((((...(((((((......((((((((..(((((((...((((((....))))))..))))).)).....))))))))...)))))))..)))))))..... ( -40.80) >DroAna_CAF1 38185 114 - 1 AAUUAAACCGAAC----CAAAAAGCGUUUACGGCCAUGGGUGACUCCCUGGAGAACACCGACACCUCGGUGGAUCCGGGCGUGAAUGUGUCUAUAGCCAAUUACGAUGCAUUUGCAAA .............----......(((((((((.((..(((.....))).(((...((((((....))))))..))))).)))))))))..................(((....))).. ( -36.00) >DroPer_CAF1 72844 111 - 1 AAUUUUG---UAU----UUAAAACCAUUGGUGGACAUGGGUGAUUCCCUGGAAAACACCGACACAUCGGUGGAUCCCGGCGUGAAUGUGUCAAUAGCCAACUACGAUGCAUUUGCAAA .......---...----......(((((((((....(.((((.(((....)))..)))).)..)))))))))........(..((((((((..(((....))).))))))))..)... ( -30.50) >consensus AAUUUAA___UAA____CAAAAACCGUUGAAGGACAUGGGUGAUUCCCUGGAAAACACCGACACAUCGGUGGAUCCAGGCGUGAAUGUGUCAAUAGCCAACUACGAUGCAUUUGCAAA ..............................................((((((...((((((....))))))..)))))).((((((.((((.............)))).))))))... (-19.05 = -19.39 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:07 2006