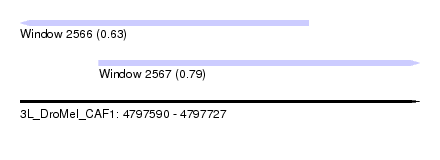
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,797,590 – 4,797,727 |
| Length | 137 |
| Max. P | 0.792384 |

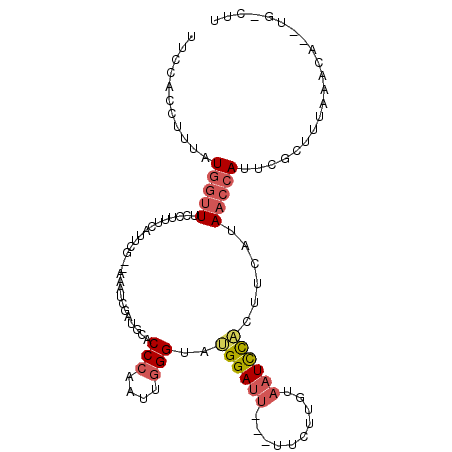
| Location | 4,797,590 – 4,797,689 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -15.33 |
| Consensus MFE | -4.72 |
| Energy contribution | -5.66 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4797590 99 - 23771897 UUCCACCUUUCUGGUUUCCUUUCAUUCG--AAAUCGAUGCACCCAAUUGGGUAUGGAUU---UUCUUAUAAUCCACUUCAUAACCAUUCGCUUUAAACA--UG-CUU ...........(((((.........(((--....)))...((((....)))).((((((---.......))))))......)))))...((........--.)-).. ( -16.10) >DroSec_CAF1 24616 99 - 1 UUUUACCUUUAUGGUUUCCUUUCAUCCG--UAAUCGAUGCACCCAAUUGGGUAAGGAUU---UUAUUAUAAUCCACUUCAUAACCAUUCGCUUUAAACA--UG-CUU ..........((((((......((((..--.....)))).((((....))))..(((((---.......))))).......))))))..((........--.)-).. ( -15.80) >DroEre_CAF1 26394 98 - 1 -UCCUCCUUUAUGGUUUCCUUUCAUUCG--AAAUCGAUGCACCCGAUUGGGCAUGGAUU---UUCUUGUAAUCCAAUUCAUAACCAUUCGCUUUAAACA--UG-CUU -.......((((((........((...(--(((((.((((.((.....)))))).))))---))..)).........))))))......((........--.)-).. ( -16.03) >DroWil_CAF1 26130 94 - 1 UUCUACUUGUUUA-AUGCCUUUCCUUUGGGCAAUC--------UUAU----UCAGGAAUCUAUUCAUGUUAUUUUCUUCAUAAUCAUCCACUUUAAACACAUGUCUA .......((((((-((((((.......)))))...--------((((----..(((((..((.......))..))))).)))).........)))))))........ ( -11.80) >DroYak_CAF1 24984 99 - 1 UUCCGCCUUUAUGGUUUCCCUUCAUUCA--AAAUCGAUGCACCCAAUUGGGCAUGGAUU---UUCUUGUAAUCCACUUCAUAACCAUUUGCUUUAAACA--UG-CUU ....((....((((((......((...(--(((((.((((.((.....)))))).))))---))..)).............))))))..))........--..-... ( -16.91) >consensus UUCCACCUUUAUGGUUUCCUUUCAUUCG__AAAUCGAUGCACCCAAUUGGGUAUGGAUU___UUCUUGUAAUCCACUUCAUAACCAUUCGCUUUAAACA__UG_CUU ...........(((((.........................(((....)))..((((((..........))))))......)))))..................... ( -4.72 = -5.66 + 0.94)


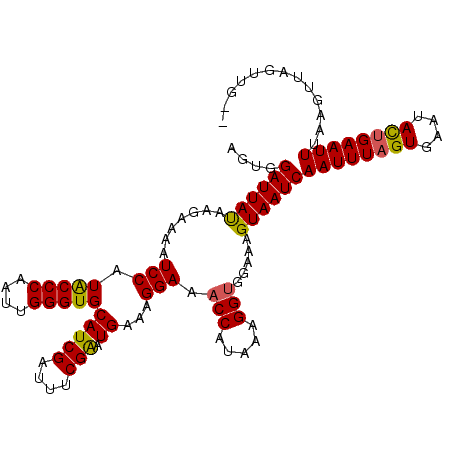
| Location | 4,797,617 – 4,797,727 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4797617 110 + 23771897 AGUGGAUUAUAAGAAAAUCCAUACCCAAUUGGGUGCAUCGAUUUCGAAUGAAAGGAAACCAGAAAGGUGGAAAGUAAUCAAUUUAGUGAAUAUAGAAUUUAAGUUAGUUA-- ....((((((.......(((.(((((....)))))(((((....)).)))...))).(((.....))).....))))))(((((((............))))))).....-- ( -19.10) >DroSec_CAF1 24643 110 + 1 AGUGGAUUAUAAUAAAAUCCUUACCCAAUUGGGUGCAUCGAUUACGGAUGAAAGGAAACCAUAAAGGUAAAAAGUAAUCAAUUUAGUGAAUACUGAAUUUAAGUUAGUUG-- ....((((((.......(((((((((....)))).((((.......)))).))))).(((.....))).....))))))((((((((....))))))))...........-- ( -23.80) >DroEre_CAF1 26421 111 + 1 AUUGGAUUACAAGAAAAUCCAUGCCCAAUCGGGUGCAUCGAUUUCGAAUGAAAGGAAACCAUAAAGGAGGA-AGUAAUCAAUUUAGUGAAUACUGAAUUUAAAUUGACUGGA ....((((((.......(((..((((....)))).(((((....)).)))...)))..((.....))....-.))))))((((((((....))))))))............. ( -22.70) >consensus AGUGGAUUAUAAGAAAAUCCAUACCCAAUUGGGUGCAUCGAUUUCGAAUGAAAGGAAACCAUAAAGGUGGAAAGUAAUCAAUUUAGUGAAUACUGAAUUUAAGUUAGUUG__ ....((((((.......(((.(((((....)))))(((((....))).))...))).(((.....))).....))))))((((((((....))))))))............. (-19.91 = -19.70 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:06 2006