| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,793,024 – 4,793,138 |
| Length | 114 |
| Max. P | 0.944045 |

| Location | 4,793,024 – 4,793,138 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4793024 114 - 23771897 AACCGA-----U-CAAUCAAUCAACUAAUCGACGAUGUGCAGCGGUCAAGUCAAUCCGCAAGCAACAGCUCGUUGAGGUGCGAACCAUGGCUAAUCCGCCAUCGGUGGUCAAAUUGGCUC .(((((-----.-...............((((((((((((.((((..........))))..)).)))..)))))))(((....))).((((......)))))))))((((.....)))). ( -33.30) >DroVir_CAF1 29201 100 - 1 CG-----------CAAUCAAACAACAAA---------CACAGCCGUGAAGUCCAUACGCAAACAGCAGCUCGUUGAGGUGCGAACGAUGGCGAAUCCGCCGUCCGUGGUGAAAUUGGCAC ..-----------...............---------....((((......((((.((((..((((.....))))...))))...(((((((....))))))).))))......)))).. ( -30.70) >DroPse_CAF1 55490 96 - 1 AACCAA-----U-CAAUCAAUC------------------AGCCGUCAAAUCGAUACGCAAGCAACAGCUCGUUGAAGUGCGAACCAUGGCCAAUCCACCGUCGGUGGUCAAAUUGGCCC ......-----.-.........------------------....(((.....))).((((..((((.....))))...))))......((((((((((((...)))))....))))))). ( -26.00) >DroMoj_CAF1 53849 109 - 1 CACCAAACCAACCCAAUUGAACCACCAA-----------UAGCUGUGAAGUCUAUACGCAAACAGCAACUCGUUGAGGUGCGAACGAUGGCGAAUCCACCGUCGGUGGUUAAAUUGGCAC ............((((((.(((((((.(-----------(((((....)).)))).((((..((((.....))))...))))...(((((.(....).)))))))))))).))))))... ( -36.30) >DroAna_CAF1 24777 110 - 1 AACCGA-----C-CAACCAAUC----CAUCGACGAUGUGCAGCGGUCAAGUCGAUACGCAAGCAACAGCUCGUUGAGGUGCGAACCAUGGCUAAUCCACCAUCGGUGGUCAAAUUGGCUC ....((-----(-((.((....----..((((((((((((.((((((.....))).)))..)).)))..)))))))((((.((...........))))))...))))))).......... ( -31.90) >DroPer_CAF1 59053 96 - 1 AACCAA-----U-CAAUCAAUC------------------AGCCGUCAAAUCGAUACGCAAGCAACAGCUCGUUGAAGUGCGAACCAUGGCCAAUCCACCGUCGGUGGUCAAAUUGGCCC ......-----.-.........------------------....(((.....))).((((..((((.....))))...))))......((((((((((((...)))))....))))))). ( -26.00) >consensus AACCAA_____U_CAAUCAAUC____AA___________CAGCCGUCAAGUCGAUACGCAAGCAACAGCUCGUUGAGGUGCGAACCAUGGCCAAUCCACCGUCGGUGGUCAAAUUGGCAC ........................................................((((..((((.....))))...))))......(((((((((((.....))))....))))))). (-19.80 = -19.80 + 0.00)


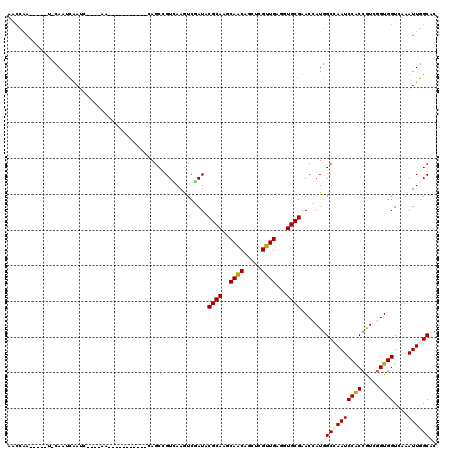
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:04 2006