| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,770,419 – 4,770,535 |
| Length | 116 |
| Max. P | 0.894689 |

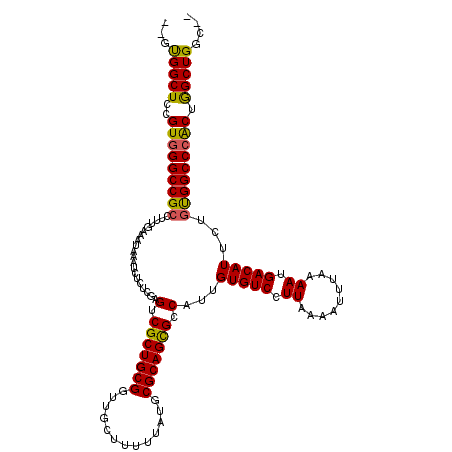
| Location | 4,770,419 – 4,770,535 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -31.29 |
| Energy contribution | -31.99 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4770419 116 + 23771897 --GCCAGCUAGUGGGCCACAGAAUGUCAUUUUAAAUUUUAAGGACACAAUGGCGCUGCGCAUAAAAAGCAACCGCAGCGACUCGAGAGAUUUAUUUCAAAGGCGGCCCACGGAGCCAC-- --....(((.(((((((.((...((((.(((........)))))))...))((((((((.............)))))).....((((......))))....)))))))))..)))...-- ( -37.12) >DroSec_CAF1 33176 116 + 1 --GCCAGCCAGUGGGCCACAGAAUGUCAUUUUAAAUUUUAAGGACACAAUGGCGCUGCGCAUAAAAAGCAACCGCAGCGACUCGAGAGAUUUAUUUCAAAGGCGGCCCACGGAGCCAC-- --((...((.(((((((.((...((((.(((........)))))))...))((((((((.............)))))).....((((......))))....))))))))))).))...-- ( -36.32) >DroSim_CAF1 32115 116 + 1 --GCCAGCCAGUGGGCCACAGAAUGUCAUUUUAAAUUUUAAGGACACAAUGGCGCUGCGCAUAAAAAGCAACCGCAGCGACUCGAGAGAUUUAUUUCAAAGGCGGCCCACGGAGCCAC-- --((...((.(((((((.((...((((.(((........)))))))...))((((((((.............)))))).....((((......))))....))))))))))).))...-- ( -36.32) >DroEre_CAF1 22470 116 + 1 --GCCAGCCAGUGGGCCACAGAAUGUCAUUUUAAAUUUUAAGGACACAAUGGCGCUGCGCAUAAAAAGCAACCGCAGCGACUCGAGAGAUUUAUUUCAAAGGCGGCCCACGGAGCCAC-- --((...((.(((((((.((...((((.(((........)))))))...))((((((((.............)))))).....((((......))))....))))))))))).))...-- ( -36.32) >DroYak_CAF1 29663 116 + 1 --GGCAGCCAGUGGGCCACAGAAUGUCAUUUUAAAUUUUAAGGACACAAUGGCGCUGCGCAUAAAAAGCAACCGCAGCGACUCGAGAGAUUUAUUUCAAAGGCGGCCCACGGAGCCAC-- --(((..((.(((((((.((...((((.(((........)))))))...))((((((((.............)))))).....((((......))))....))))))))))).)))..-- ( -40.02) >DroAna_CAF1 23383 120 + 1 ACUCCAGCCAGCGGGCCGGCGAAUGUCAUUUUAAAUUUUAAGGACACAAUGGCACUGCGCAUAAAAAGCAACCGCAGCGACUCGAGAGAUUUAUUUCAAAGGCGGCCAACGGAGCCGCCU ......((..((((((((.....((((.(((........)))))))...))))..(((.........))).)))).)).....((((......))))..((((((((....).))))))) ( -34.10) >consensus __GCCAGCCAGUGGGCCACAGAAUGUCAUUUUAAAUUUUAAGGACACAAUGGCGCUGCGCAUAAAAAGCAACCGCAGCGACUCGAGAGAUUUAUUUCAAAGGCGGCCCACGGAGCCAC__ ......(((.(((((((.((...((((.(((........)))))))...))((((((((.............)))))).....((((......))))....))))))))).).))..... (-31.29 = -31.99 + 0.70)

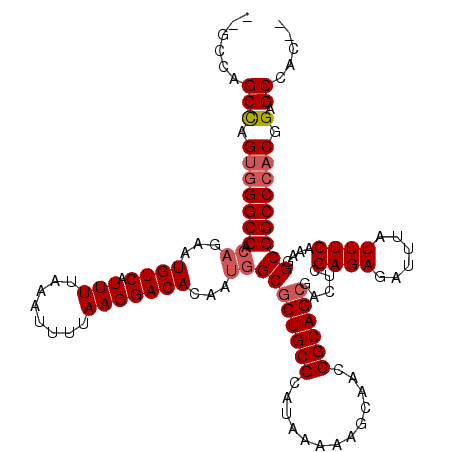
| Location | 4,770,419 – 4,770,535 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.02 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -35.35 |
| Energy contribution | -34.99 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4770419 116 - 23771897 --GUGGCUCCGUGGGCCGCCUUUGAAAUAAAUCUCUCGAGUCGCUGCGGUUGCUUUUUAUGCGCAGCGCCAUUGUGUCCUUAAAAUUUAAAAUGACAUUCUGUGGCCCACUAGCUGGC-- --..((((..(((((((((....................(.(((((((.............))))))).)...(((((.((.........)).)))))...))))))))).))))...-- ( -36.42) >DroSec_CAF1 33176 116 - 1 --GUGGCUCCGUGGGCCGCCUUUGAAAUAAAUCUCUCGAGUCGCUGCGGUUGCUUUUUAUGCGCAGCGCCAUUGUGUCCUUAAAAUUUAAAAUGACAUUCUGUGGCCCACUGGCUGGC-- --..((((..(((((((((....................(.(((((((.............))))))).)...(((((.((.........)).)))))...))))))))).))))...-- ( -36.42) >DroSim_CAF1 32115 116 - 1 --GUGGCUCCGUGGGCCGCCUUUGAAAUAAAUCUCUCGAGUCGCUGCGGUUGCUUUUUAUGCGCAGCGCCAUUGUGUCCUUAAAAUUUAAAAUGACAUUCUGUGGCCCACUGGCUGGC-- --..((((..(((((((((....................(.(((((((.............))))))).)...(((((.((.........)).)))))...))))))))).))))...-- ( -36.42) >DroEre_CAF1 22470 116 - 1 --GUGGCUCCGUGGGCCGCCUUUGAAAUAAAUCUCUCGAGUCGCUGCGGUUGCUUUUUAUGCGCAGCGCCAUUGUGUCCUUAAAAUUUAAAAUGACAUUCUGUGGCCCACUGGCUGGC-- --..((((..(((((((((....................(.(((((((.............))))))).)...(((((.((.........)).)))))...))))))))).))))...-- ( -36.42) >DroYak_CAF1 29663 116 - 1 --GUGGCUCCGUGGGCCGCCUUUGAAAUAAAUCUCUCGAGUCGCUGCGGUUGCUUUUUAUGCGCAGCGCCAUUGUGUCCUUAAAAUUUAAAAUGACAUUCUGUGGCCCACUGGCUGCC-- --(..(((..(((((((((....................(.(((((((.............))))))).)...(((((.((.........)).)))))...))))))))).)))..).-- ( -39.12) >DroAna_CAF1 23383 120 - 1 AGGCGGCUCCGUUGGCCGCCUUUGAAAUAAAUCUCUCGAGUCGCUGCGGUUGCUUUUUAUGCGCAGUGCCAUUGUGUCCUUAAAAUUUAAAAUGACAUUCGCCGGCCCGCUGGCUGGAGU ((((((((.....)))))))).............(((.(((((..((((((((.........)))).(((...(((((.((.........)).))))).....)))))))))))).))). ( -37.40) >consensus __GUGGCUCCGUGGGCCGCCUUUGAAAUAAAUCUCUCGAGUCGCUGCGGUUGCUUUUUAUGCGCAGCGCCAUUGUGUCCUUAAAAUUUAAAAUGACAUUCUGUGGCCCACUGGCUGGC__ ...(((((..(((((((((....................(.(((((((.............))))))).)...(((((.((.........)).)))))...))))))))).))))).... (-35.35 = -34.99 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:50 2006