
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,768,736 – 4,768,975 |
| Length | 239 |
| Max. P | 0.973059 |

| Location | 4,768,736 – 4,768,826 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -31.02 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4768736 90 - 23771897 ---------GCAUUGGGCUUGUAAAUGGGCUUUUAUUUUCGUUUGCCGGGUUAAGAGCCCGCACAAGGACGAAUGCCACCGCGACCA-AGACGCGGACGU ---------(((((.(.(((((...((((((((((..((((.....)))).)))))))))).)))))..).)))))..(((((....-...))))).... ( -34.70) >DroSec_CAF1 31519 90 - 1 ---------GCAGUGGGCUUGUAAAUGGGCUUUUAUUUUCGUUGGCCGGGUUAAGAGCCCGCACAAGGACGAAUGCCACCGCGGCCA-AGACGCGGACGC ---------(((.((..(((((...((((((((((..((((.....)))).)))))))))).)))))..))..)))..(((((....-...))))).... ( -32.90) >DroSim_CAF1 30488 90 - 1 ---------GCAGUGGGCUUGUAAAUGGGCUUUUAUUUUCGUUGGCCGGGUUAAGAGCCUGCACAAGGACGAAUGCCACCGCGGCCA-AGACGCGGACGU ---------(((.((..(((((...((((((((((..((((.....)))).)))))))))).)))))..))..)))..(((((....-...))))).... ( -30.00) >DroEre_CAF1 20780 91 - 1 ---------GCAGUGGGCUUGUAAAUGGGCUUUUAUUUUCGUUGGCCGGGUUAAGAGCCCGCACAAGGACGAAUGCCACCGCGGCCAAAGACGCGGACGU ---------(((.((..(((((...((((((((((..((((.....)))).)))))))))).)))))..))..)))..(((((........))))).... ( -33.20) >DroYak_CAF1 27906 99 - 1 CUGGCAGUGGCAGUGGGCUUGUAAAUGGGCUUUUAUUUUCGUUAGCCGGGUUAAGAGCCGGCACAAGGACGAAUGCCACCGCGGCCA-AGACGCGGACGU .......(((((..((((((......)))))).....((((((.(((((.(....).))))).....)))))))))))(((((....-...))))).... ( -34.60) >consensus _________GCAGUGGGCUUGUAAAUGGGCUUUUAUUUUCGUUGGCCGGGUUAAGAGCCCGCACAAGGACGAAUGCCACCGCGGCCA_AGACGCGGACGU .........(((..(..(((((...((((((((((..((((.....)))).)))))))))).)))))..)...)))..(((((........))))).... (-31.02 = -31.10 + 0.08)



| Location | 4,768,826 – 4,768,935 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -29.78 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4768826 109 + 23771897 ---CACAGUCAAGGACUUGAGCUUG------AGCCUGAGCCGGCUGGCAAUUCAAAUGCCAGGACACGAGGACA--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUC ---.((.((((.(..((((.((((.------.....))))...((((((.......))))))....))))..).--))))..(((((((.((((...)))).)))).)))...))..... ( -35.60) >DroSec_CAF1 31609 109 + 1 ---CACAGUCAAGGACUUGAGCUUG------AGCCUGAGCCGGCUGGCAAUUCAAAUGCCAGGACACAAGGACA--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUC ---.((.((((.(..((((.((((.------.....))))...((((((.......))))))....))))..).--))))..(((((((.((((...)))).)))).)))...))..... ( -35.90) >DroSim_CAF1 30578 109 + 1 ---CACAGUCAAGGACUUGAGCUUG------AGCCUGAGCCGGCUGGCAAUUCAAAUGCCAGGACACGAGGACA--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUC ---.((.((((.(..((((.((((.------.....))))...((((((.......))))))....))))..).--))))..(((((((.((((...)))).)))).)))...))..... ( -35.60) >DroEre_CAF1 20871 115 + 1 ---CACAGUCAAGGACUUCAGCUUGAGCCUGAGCCUGAGCCGGCUGGCAAUUCAAAUGCCAGGACACGAGGACU--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUC ---.((.((((((..((((.((((..((....))..))))...((((((.......)))))).....)))).))--))))..(((((((.((((...)))).)))).)))...))..... ( -39.90) >DroYak_CAF1 28005 112 + 1 UGCCACAGUCAAGGACUUGAGCUUG------AGCCUGAGCCGGCUGGCAAUUCAAAUGCCAGGACACAAGGACA--UGGCAGGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUC (((((..(((..((.(((..((...------.))..)))))..((((((.......))))))........))).--))))).(((((((.((((...)))).)))).))).......... ( -38.20) >DroAna_CAF1 21796 110 + 1 UGCCACAGUCAAGGACUUGAGCC---------G-CUUGGCAGCCUAGCAAUUCAAAUGCCAAGAGACGAGGACAGCCGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUC (((.........(..((((....---------.-(((((((....(....).....)))))))...))))..).((((..(((((((....)))...))))..)))).)))......... ( -34.50) >consensus ___CACAGUCAAGGACUUGAGCUUG______AGCCUGAGCCGGCUGGCAAUUCAAAUGCCAGGACACGAGGACA__UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUC ....((.((((.(..((((.((((............))))...((((((.......))))))....))))..)...))))..(((((((.((((...)))).)))).)))...))..... (-29.78 = -30.12 + 0.34)

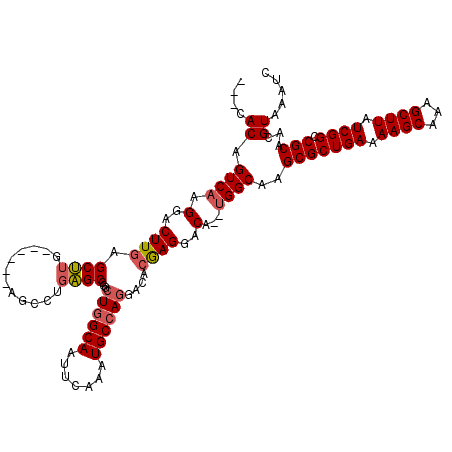

| Location | 4,768,857 – 4,768,975 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.46 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -26.29 |
| Energy contribution | -26.48 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
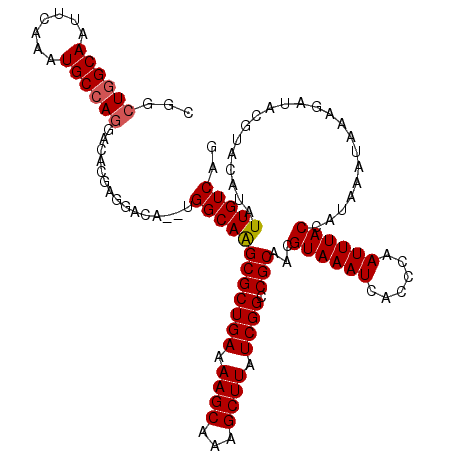
>3L_DroMel_CAF1 4768857 118 + 23771897 CGGCUGGCAAUUCAAAUGCCAGGACACGAGGACA--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUCACCCAAUUUACCAUAAAUAAAGAUACGUACAUAUUGUCAG ...((((((((.....((((((..(....)..).--))))).(((((((.((((...)))).)))).)))...((((((......)))))).....................)))))))) ( -30.10) >DroSec_CAF1 31640 118 + 1 CGGCUGGCAAUUCAAAUGCCAGGACACAAGGACA--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUCACCCAAUUUACCAUAAAUAAAGAUAUGUACAUAUUGUCAG ...((((((.......))))))((((....((((--((.(..(((((((.((((...)))).)))).)))...((((((......))))))..........).))))).)....)))).. ( -31.10) >DroSim_CAF1 30609 118 + 1 CGGCUGGCAAUUCAAAUGCCAGGACACGAGGACA--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUCACCCAAUUUACCAUAAAUAAAGAUACGUACAUAUUGUCAG ...((((((((.....((((((..(....)..).--))))).(((((((.((((...)))).)))).)))...((((((......)))))).....................)))))))) ( -30.10) >DroEre_CAF1 20908 118 + 1 CGGCUGGCAAUUCAAAUGCCAGGACACGAGGACU--UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUCACCCAAUUUACCAUAAAUAAAGAUACGUACAUAUUGUCAG ...((((((((.....(((((((.(.....).))--))))).(((((((.((((...)))).)))).)))...((((((......)))))).....................)))))))) ( -31.00) >DroYak_CAF1 28039 118 + 1 CGGCUGGCAAUUCAAAUGCCAGGACACAAGGACA--UGGCAGGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUCACCCAAUUUACCAUAAAUAAAGAUACGUACAUAUUGUCAG ...((((((((.....((((((..(....)..).--))))).(((((((.((((...)))).)))).)))...((((((......)))))).....................)))))))) ( -29.90) >DroAna_CAF1 21826 120 + 1 AGCCUAGCAAUUCAAAUGCCAAGAGACGAGGACAGCCGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUCACCCAAUUUACCAUAAAUAAAGAUACGUACAUAUUGUCAG ......(((.......))).....(((((.(((.((((..(((((((....)))...))))..))))......((((((......))))))...............)).)...))))).. ( -26.10) >consensus CGGCUGGCAAUUCAAAUGCCAGGACACGAGGACA__UGGCAAGCGCUGAAAAGCAAAGCUUAUCGGCCGCAACGUAAAUCACCCAAUUUACCAUAAAUAAAGAUACGUACAUAUUGUCAG ...((((((.......))))))...............((((((((((((.((((...)))).)))).)))...((((((......))))))......................))))).. (-26.29 = -26.48 + 0.19)

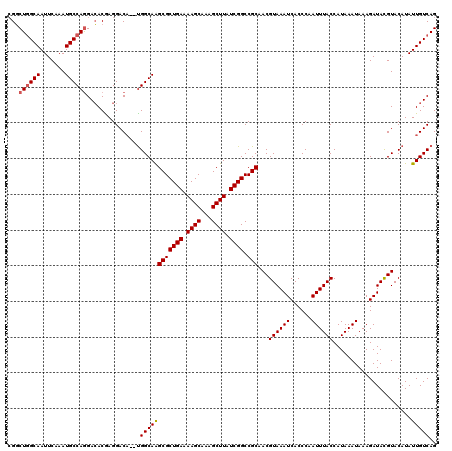
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:47 2006