
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,766,226 – 4,766,323 |
| Length | 97 |
| Max. P | 0.984084 |

| Location | 4,766,226 – 4,766,323 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
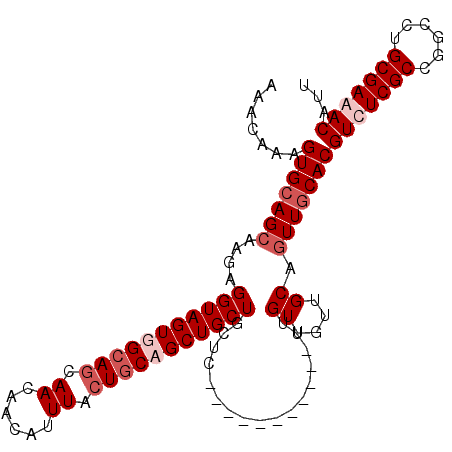
>3L_DroMel_CAF1 4766226 97 + 23771897 AAUGUUUCGCAGGCCGGCGAAACGUGCAACUGCAACAACAA-----------AAGUAGCAGCUGUAGUAAAUGUUGUUGCUGCCACUACCUCUUGCUGCACUUUGUUU ...(((((((......)))))))(((((...((((......-----------..((((((((..((.....))..)))))))).........)))))))))....... ( -29.73) >DroSec_CAF1 29027 97 + 1 AAUGUUUCGCAGGCCGGCGAGACGUGCAACUGCAACAACAA-----------GAGCAGCAGCUGCAGUAAAUGUUGUUGCUGCAACUACCUCUUGCUGCACUUUGUUU ...(((((((......)))))))(((((...((((......-----------..((((((((.(((.....))).)))))))).........)))))))))....... ( -34.93) >DroSim_CAF1 27975 97 + 1 AAUGUUUCGCAGGCCGGCGAGACGUGCAACUGCAACAACAA-----------GAGCAGCAGCUGCAGUAAAUGUUGUUGCUGCAACUACCUCUUUCUGCACUUUGUUU ...(((((((......)))))))(((((.((.........)-----------).((((((((.(((.....))).)))))))).............)))))....... ( -32.90) >DroYak_CAF1 25274 107 + 1 AAUGUUUCGCAGGCCGGCGAAACGUGCAACUGCAGCAACU-GCAACAACAGAAAGCAGCAGCUGCAUUAAACGUUGUUGCUGCCACUACCUAUUGCUUCACUUUGUUU .(((((((((......)))))))))((((.((.(((((((-(......)))...((((((((.((.......)).)))))))).........))))).))..)))).. ( -34.00) >consensus AAUGUUUCGCAGGCCGGCGAAACGUGCAACUGCAACAACAA___________AAGCAGCAGCUGCAGUAAAUGUUGUUGCUGCAACUACCUCUUGCUGCACUUUGUUU .(((((((((......)))))))))((....)).....................((((((((..((.....))..))))))))......................... (-27.50 = -27.12 + -0.38)



| Location | 4,766,226 – 4,766,323 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -27.85 |
| Energy contribution | -29.85 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4766226 97 - 23771897 AAACAAAGUGCAGCAAGAGGUAGUGGCAGCAACAACAUUUACUACAGCUGCUACUU-----------UUGUUGUUGCAGUUGCACGUUUCGCCGGCCUGCGAAACAUU .....((.((((((((.(((.(((((((((................))))))))))-----------)).)))))))).))....(((((((......)))))))... ( -36.99) >DroSec_CAF1 29027 97 - 1 AAACAAAGUGCAGCAAGAGGUAGUUGCAGCAACAACAUUUACUGCAGCUGCUGCUC-----------UUGUUGUUGCAGUUGCACGUCUCGCCGGCCUGCGAAACAUU .......((((((((((((((((((((((.((......)).))))))))))..)))-----------))((....)).)))))))((.((((......)))).))... ( -35.80) >DroSim_CAF1 27975 97 - 1 AAACAAAGUGCAGAAAGAGGUAGUUGCAGCAACAACAUUUACUGCAGCUGCUGCUC-----------UUGUUGUUGCAGUUGCACGUCUCGCCGGCCUGCGAAACAUU .......((((((.(((((((((((((((.((......)).))))))))))..)))-----------))((....))..))))))((.((((......)))).))... ( -31.90) >DroYak_CAF1 25274 107 - 1 AAACAAAGUGAAGCAAUAGGUAGUGGCAGCAACAACGUUUAAUGCAGCUGCUGCUUUCUGUUGUUGC-AGUUGCUGCAGUUGCACGUUUCGCCGGCCUGCGAAACAUU ............(((((..((((..((.(((((((((......((((...))))....)))))))))-.))..)))).)))))..(((((((......)))))))... ( -41.40) >consensus AAACAAAGUGCAGCAAGAGGUAGUGGCAGCAACAACAUUUACUGCAGCUGCUGCUC___________UUGUUGUUGCAGUUGCACGUCUCGCCGGCCUGCGAAACAUU .......(((((((....(((((((((((.((......)).))))))))))).................((....)).)))))))(((((((......)))))))... (-27.85 = -29.85 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:42 2006