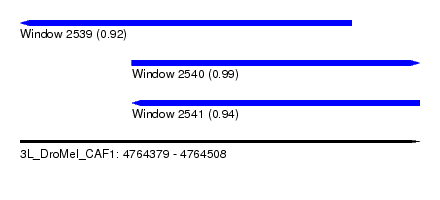
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,764,379 – 4,764,508 |
| Length | 129 |
| Max. P | 0.988601 |

| Location | 4,764,379 – 4,764,486 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -20.92 |
| Energy contribution | -20.36 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
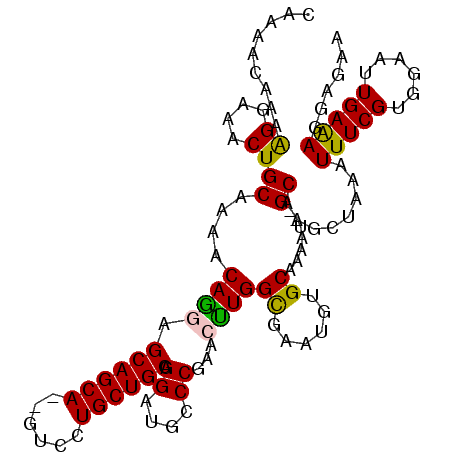
>3L_DroMel_CAF1 4764379 107 - 23771897 CAAAACAAAAGGAAACUGCAAAACAGGAGCAGCA--GUCCUGCUGCAGGAUGCCCGAACUUGGUGAAUGUGCAAAAU--GCAAGCUAAAUUUCGUGGAAUUGAAAGGAGAA .........((....))...........((((((--....))))))..(((.((((((.(((((.....(((.....--))).)))))..)))).)).))).......... ( -24.70) >DroSec_CAF1 27142 107 - 1 CAAAACAAAAGGAAACUGCAAAACAGGAGCAGCA--GUCCUGCUGCAGGAUGCCCGAACCUGGCGAAUGUGCAAAAU--GCAAGCUAAAUUUCGUGGAAUUGAAAGGAGAA .........((....))(((...((((.((((((--....)))))).((....))...))))(((....)))....)--))...((...(((((......)))))..)).. ( -28.00) >DroSim_CAF1 24436 107 - 1 CAAAACAAAAGCAAACUGCAAAACAGGAGCAGCA--GUCCUGCUGCAGGAUGCCCGAACCUGGCGAAUGGGCAAAAU--GCAAGCUAAAUUUCGUGGAAUUGAAAGGAGAA .........(((....((((...((((.((((((--....)))))).((....))...))))((......))....)--))).)))...(((((......)))))...... ( -26.80) >DroEre_CAF1 16352 109 - 1 CAAAACAAAGGGAAACUGCAAAACAAAAGCAGCA--GUCCUGCUGCAGGAUGCCCGAAAUUGGCGAAUGUGCAAAAUGUGCAAGGUAAAUUUCGUGGAAUUGAGAGGAGAA .........((....))...........((((((--....))))))..(((.(((((((((.((...((..(.....)..))..)).))))))).)).))).......... ( -27.30) >DroYak_CAF1 23362 109 - 1 CAAAACAAAAGGAAACUGCAAAACAAAAGCAGCAGAGUCCUGCUGCAGGAUGCCCAAAAUUGGCGAGUGUGCAAAAU--GCAGGUUAAAUUUCGUGGAAUUGAAAGGAGAA ...............(((((........(((((((....)))))))....((((.......))))...........)--))))......(((((......)))))...... ( -26.00) >consensus CAAAACAAAAGGAAACUGCAAAACAGGAGCAGCA__GUCCUGCUGCAGGAUGCCCGAACUUGGCGAAUGUGCAAAAU__GCAAGCUAAAUUUCGUGGAAUUGAAAGGAGAA .........((....))((....((((.((((((......)))))).((....))...))))((......)).......))........(((((......)))))...... (-20.92 = -20.36 + -0.56)

| Location | 4,764,415 – 4,764,508 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
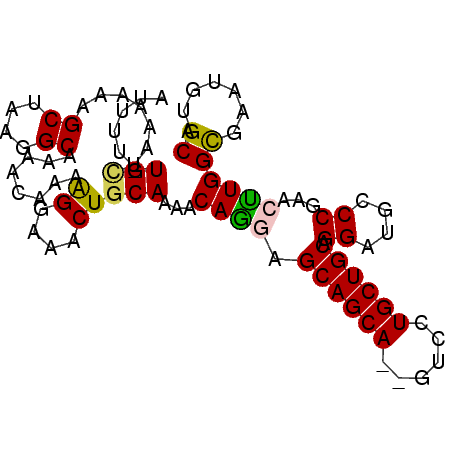
>3L_DroMel_CAF1 4764415 93 + 23771897 UGCACAUUCACCAAGUUCGGGCAUCCUGCAGCAGGAC--UGCUGCUCCUGUUUUGCAGUUUCCUUUUGUUUUGCCUUAGCUUUUUUAGCA-AAAAU (((.........(((((.(((((....((((.((((.--.(((((.........))))).)))).))))..))))).))))).....)))-..... ( -26.74) >DroSec_CAF1 27178 93 + 1 UGCACAUUCGCCAGGUUCGGGCAUCCUGCAGCAGGAC--UGCUGCUCCUGUUUUGCAGUUUCCUUUUGUUUUGCCUUAGCUUUUUUAGCA-AAAAU .........((.(((((.(((((....((((.((((.--.(((((.........))))).)))).))))..))))).))))).....)).-..... ( -26.90) >DroSim_CAF1 24472 93 + 1 UGCCCAUUCGCCAGGUUCGGGCAUCCUGCAGCAGGAC--UGCUGCUCCUGUUUUGCAGUUUGCUUUUGUUUUGCUUUAGCUUUUUUAGCA-AAAAU (((((..((....))...)))))..((((((((((((--....).))))))..)))))((((((........((....))......))))-))... ( -28.24) >DroEre_CAF1 16390 93 + 1 UGCACAUUCGCCAAUUUCGGGCAUCCUGCAGCAGGAC--UGCUGCUUUUGUUUUGCAGUUUCCCUUUGUUUUGCCAUAGCUUUUUUACCA-AAAAU .((......))........((((....((((..(((.--.(((((.........))))).)))..))))..))))...............-..... ( -20.00) >DroYak_CAF1 23398 96 + 1 UGCACACUCGCCAAUUUUGGGCAUCCUGCAGCAGGACUCUGCUGCUUUUGUUUUGCAGUUUCCUUUUGUUUUGCCUUAGCUUUUUUAACAAAAAAU ((((.....(((.......))).....(((((((....)))))))........))))......(((((((..((....))......)))))))... ( -23.20) >consensus UGCACAUUCGCCAAGUUCGGGCAUCCUGCAGCAGGAC__UGCUGCUCCUGUUUUGCAGUUUCCUUUUGUUUUGCCUUAGCUUUUUUAGCA_AAAAU ............(((((.(((((....((((.((((....(((((.........))))).)))).))))..))))).))))).............. (-20.10 = -20.70 + 0.60)



| Location | 4,764,415 – 4,764,508 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -19.00 |
| Energy contribution | -18.68 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4764415 93 - 23771897 AUUUU-UGCUAAAAAAGCUAAGGCAAAACAAAAGGAAACUGCAAAACAGGAGCAGCA--GUCCUGCUGCAGGAUGCCCGAACUUGGUGAAUGUGCA ..(((-((((...........)))))))....((....))(((...((((.((((((--....)))))).((....))...)))).......))). ( -24.50) >DroSec_CAF1 27178 93 - 1 AUUUU-UGCUAAAAAAGCUAAGGCAAAACAAAAGGAAACUGCAAAACAGGAGCAGCA--GUCCUGCUGCAGGAUGCCCGAACCUGGCGAAUGUGCA ..(((-(((.......((....))........((....))))))))((((.((((((--....)))))).((....))...))))(((....))). ( -28.10) >DroSim_CAF1 24472 93 - 1 AUUUU-UGCUAAAAAAGCUAAAGCAAAACAAAAGCAAACUGCAAAACAGGAGCAGCA--GUCCUGCUGCAGGAUGCCCGAACCUGGCGAAUGGGCA .....-..........(((...((.........((.....))....((((.((((((--....)))))).((....))...)))))).....))). ( -25.60) >DroEre_CAF1 16390 93 - 1 AUUUU-UGGUAAAAAAGCUAUGGCAAAACAAAGGGAAACUGCAAAACAAAAGCAGCA--GUCCUGCUGCAGGAUGCCCGAAAUUGGCGAAUGUGCA .....-..(((.....((((.((((.......((....))...........((((((--....))))))....))))......)))).....))). ( -23.80) >DroYak_CAF1 23398 96 - 1 AUUUUUUGUUAAAAAAGCUAAGGCAAAACAAAAGGAAACUGCAAAACAAAAGCAGCAGAGUCCUGCUGCAGGAUGCCCAAAAUUGGCGAGUGUGCA ......(((.......(((((((((.......((....))...........(((((((....)))))))....)))).....)))))......))) ( -25.22) >consensus AUUUU_UGCUAAAAAAGCUAAGGCAAAACAAAAGGAAACUGCAAAACAGGAGCAGCA__GUCCUGCUGCAGGAUGCCCGAACUUGGCGAAUGUGCA ......(((.......((....))........((....)))))...((((.((((((......)))))).((....))...))))((......)). (-19.00 = -18.68 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:39 2006