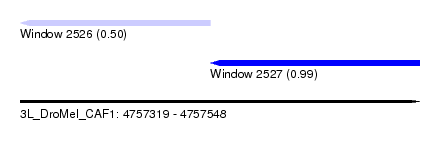
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,757,319 – 4,757,548 |
| Length | 229 |
| Max. P | 0.988595 |

| Location | 4,757,319 – 4,757,428 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.37 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4757319 109 - 23771897 CGAGUCUCCUGACUUUUUUUUCACUUUGC---------UCGCCUUUGUUGCCGAAUCA--AAGUGACUUUUGCUUGAAGUCGCCGACGACAACACAACAUUUUGUAAUUUUCGUCAUAUU .(((((....))))).......((((((.---------(((.(......).)))..))--))))((((((.....))))))...(((((....((((....)))).....)))))..... ( -23.70) >DroSec_CAF1 20024 108 - 1 CGAGUCUCCAGGCUUU-UCUUCACUUUGC---------UCGCCUUUGUGGCCGAAACA--AAGUGACUUUUGCUUGAAGUCGCCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU ((((((..(((((...-...((((((((.---------(((((.....)).)))..))--)))))).....)))))..((((....)))))))((((....)))).....)))....... ( -29.60) >DroSim_CAF1 17357 108 - 1 CGAGUCUCCUGGCUUU-UCUUCACUUUGC---------UCGCCUUUGUGGCCGAAACA--AAGUGACUUUUGCUUGAAGUCGCCGACAACGACACAACAUUUUGUAAUUUUCGUCAUAUU ((((((...((((...-.....((((((.---------(((((.....)).)))..))--))))((((((.....)))))))))).....)))((((....)))).....)))....... ( -24.60) >DroEre_CAF1 9228 108 - 1 CGAGUCUGCUGGCUUU-UCUUCACUUGGC---------UCGCCUUUGUGGCCGAAUCA--AAGUGACUUUUGCUUGAAGUCACCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU .(((((....))))).-.......(((((---------.(((....))))))))....--..((((((((.....)))))))).(((((.((.((((....))))..)).)))))..... ( -28.50) >DroYak_CAF1 15991 117 - 1 CGAGUCUCUUGGCUUU-UCUUCACUUUGUGGUCCUUUGUGGUCUUUGUGGCCGAAUCA--AAGUGACUUUUGCUUGAAGUUGCCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU (((((((((((((...-.(((((......((((((((((((((.....)))))...))--))).))))......)))))..))))).)).)))((((....)))).....)))....... ( -32.60) >DroAna_CAF1 9986 92 - 1 -----------------UUUUCACUCGCCG--------GCUCCUUUAUGGCCAAAUCAAAAAGUGACUUUUGCUUGAAGUUGCCGAC---GACACAGCUUUUUGUAAUUUUCGUCAUAUU -----------------............(--------((..(((((.(((.((((((.....))).))).))))))))..)))(((---((....((.....)).....)))))..... ( -19.40) >consensus CGAGUCUCCUGGCUUU_UCUUCACUUUGC_________UCGCCUUUGUGGCCGAAUCA__AAGUGACUUUUGCUUGAAGUCGCCGACGACGACACAACAUUUUGUAAUUUUCGUCAUAUU .(((((....))))).........................(((.....)))...........((((((((.....)))))))).(((((.((.((((....))))..)).)))))..... (-17.36 = -17.52 + 0.16)
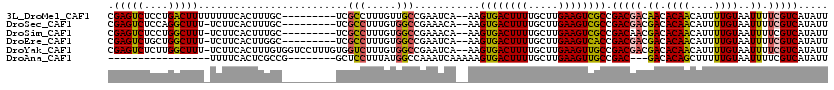
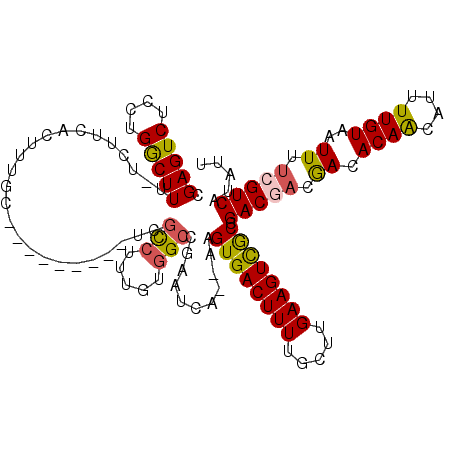
| Location | 4,757,428 – 4,757,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -36.78 |
| Energy contribution | -36.83 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4757428 120 - 23771897 GGACGGAAGUCGGUGUCAAGUGUCGCGGAAGCUUUCUGGCACUUCAUCAACUUUGAAGUAAACGUCCGCCCGCCAGGUGGAGCCACAAAUCAGAUAAGCGAGCACGCGAUUAAUAUUUUU .(((....)))..........(((((((..(((((((((.((((((.......)))))).....((((((.....))))))........))))).))))...).)))))).......... ( -38.60) >DroSec_CAF1 20132 120 - 1 GGACGGAAGUCGGUGUCAAGUGUCGCGGAAGCUUUCUGGCACUUCAUCAACUUUGAAGUAAACGUCCGCCCGCCAGGUGGAGCCACAAAUCAGAUAAGCGAGCACGCGAUUAAUAUUUUU .(((....)))..........(((((((..(((((((((.((((((.......)))))).....((((((.....))))))........))))).))))...).)))))).......... ( -38.60) >DroSim_CAF1 17465 120 - 1 GGACGGAAGUCGGUGUCAAGUGUCGCGGAAGCUUUCUGGCACUUCAUCAACUUUGAAGUAAACGUCCGCCCGCCAGGUGGAGCCACAAAUCAGAUAAGCGAGCACGCGAUUAAUAUUUUU .(((....)))..........(((((((..(((((((((.((((((.......)))))).....((((((.....))))))........))))).))))...).)))))).......... ( -38.60) >DroEre_CAF1 9336 120 - 1 GGACGGAAGUCGGUGUCAAGUGUCGCGGAAGCUUUCUGGCACUUCAUCAACUUUGAAGUAAGCGUCCGCCCGCCAGGUGGAGCCACAAAUCAGAUAAGCGAGCACGCGAUUAAUAUUUUU .(((....)))..........(((((((..(((((((((.((((((.......))))))..((.((((((.....))))))))......))))).))))...).)))))).......... ( -40.80) >DroYak_CAF1 16108 120 - 1 GGACGGAAGUCGGUGUCAAGUGUCGCGGAAGCUUUCUGGCACUUCAUCAACUUUGAAGUAAACGUCCGCCCGCCAGGUGGAGCCACAAAUCAGAUAAGGGCGCACGCGAUUAAUAUUUUU .(((....))).(((((...((((((....))(((.((((((((((.......)))))).....((((((.....)))))))))).)))...))))..)))))................. ( -41.90) >DroAna_CAF1 10078 118 - 1 GAACGGAAGUUGGUGUCAAGUGUCGCGGAAGCUUUCUGGCACUUCAUCAACUUUGAAGUAAACGUCCGCCCAAGA-AUGGAGCCACAAAUCAGAUAAGCGAGCACGCGAUUAAUAUUUU- .....((((((((((..(((((((((....)).....)))))))))))))))))((((((...(((.(((((...-.))).))..............(((....))))))...))))))- ( -31.50) >consensus GGACGGAAGUCGGUGUCAAGUGUCGCGGAAGCUUUCUGGCACUUCAUCAACUUUGAAGUAAACGUCCGCCCGCCAGGUGGAGCCACAAAUCAGAUAAGCGAGCACGCGAUUAAUAUUUUU .(((....))).((((....((((((....))(((.((((((((((.......)))))).....((((((.....)))))))))).)))...)))).....))))............... (-36.78 = -36.83 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:26 2006