
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,756,515 – 4,756,618 |
| Length | 103 |
| Max. P | 0.935647 |

| Location | 4,756,515 – 4,756,618 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.26 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
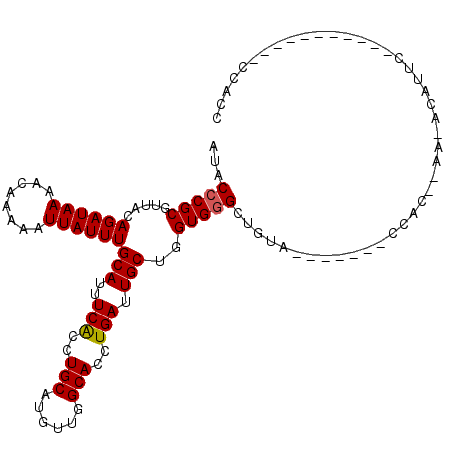
>3L_DroMel_CAF1 4756515 103 + 23771897 AUGCCCGCGUUACAGAUAAAACAAAAAUUAUUUGCAUUUCACCUGCAUGUUGGCACCUGAUUGCUGGUGGGCUGUA-GCUACCCCACUGAAAACAUUCU----------CCACC ..((((((.....((((((........))))))(((..(((..(((......)))..))).)))..))))))....-......................----------..... ( -20.40) >DroSec_CAF1 19240 80 + 1 AUACCCGCGUUACAGAUAAAACAAAAAUUAUUUGCAUUUCACCUGCAUGUUGGCACCUGAUUGCUGGUGGGCUGUA----------------------------------CACC ((((((((.....((((((........))))))(((..(((..(((......)))..))).)))..))))).))).----------------------------------.... ( -16.80) >DroSim_CAF1 16582 80 + 1 AUACCCGCGUUACAGAUAAAACAAAAAUUAUUUGCAUUUCACCUGCAUGUUGGCACCUGAUUGCUGGUGGGCUGUA----------------------------------CACC ((((((((.....((((((........))))))(((..(((..(((......)))..))).)))..))))).))).----------------------------------.... ( -16.80) >DroEre_CAF1 8467 108 + 1 AUGCACGCGUUACAGAUAAAACAAAAAUUAUUUGCAUUUCGCCUGCAAGUUGGCACCUGAUUGCUGGUGGGACGU------AACCACUAAAAACAUUCCCUAGCACCAGCCACC .((((.(((...(((((((........))))))).....))).))))...((((...((..((((((.((((.((------...........)).)))))))))).)))))).. ( -28.00) >DroYak_CAF1 15119 114 + 1 AUACCCGCGCUACAGAUAAAACAAAAAUUAUUUGCAUUUCACCUGCAUGUUGGCACCUGAUUGCUGGUGGGCUGGAAACAAACCCACAAAACACAUUCCCUAACACCAGCCAAC .((((.((....(((((((........)))))))....(((..(((......)))..)))..)).))))((((((..............................))))))... ( -23.51) >consensus AUACCCGCGUUACAGAUAAAACAAAAAUUAUUUGCAUUUCACCUGCAUGUUGGCACCUGAUUGCUGGUGGGCUGUA_______CCAC__AA_ACAUUC___________CCACC ...(((((.....((((((........))))))(((..(((..(((......)))..))).)))..)))))........................................... (-15.22 = -15.26 + 0.04)

| Location | 4,756,515 – 4,756,618 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
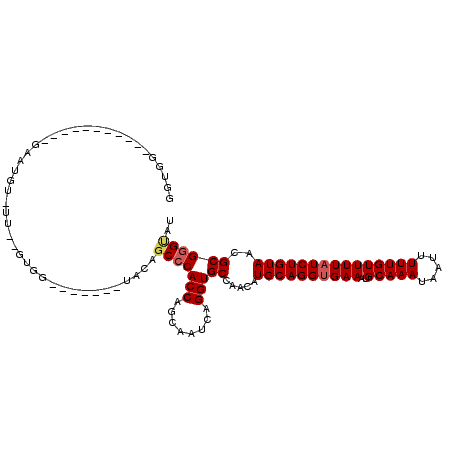
>3L_DroMel_CAF1 4756515 103 - 23771897 GGUGG----------AGAAUGUUUUCAGUGGGGUAGC-UACAGCCCACCAGCAAUCAGGUGCCAACAUGCAGGUGAAAUGCAAAUAAUUUUUGUUUUAUCUGUAACGCGGGCAU (((((----------.(((....))).((((.....)-)))...))))).........(((((..(.((((((((((..(((((.....)))))))))))))))..)..))))) ( -32.80) >DroSec_CAF1 19240 80 - 1 GGUG----------------------------------UACAGCCCACCAGCAAUCAGGUGCCAACAUGCAGGUGAAAUGCAAAUAAUUUUUGUUUUAUCUGUAACGCGGGUAU ((((----------------------------------.......)))).........(((((..(.((((((((((..(((((.....)))))))))))))))..)..))))) ( -22.10) >DroSim_CAF1 16582 80 - 1 GGUG----------------------------------UACAGCCCACCAGCAAUCAGGUGCCAACAUGCAGGUGAAAUGCAAAUAAUUUUUGUUUUAUCUGUAACGCGGGUAU ((((----------------------------------.......)))).........(((((..(.((((((((((..(((((.....)))))))))))))))..)..))))) ( -22.10) >DroEre_CAF1 8467 108 - 1 GGUGGCUGGUGCUAGGGAAUGUUUUUAGUGGUU------ACGUCCCACCAGCAAUCAGGUGCCAACUUGCAGGCGAAAUGCAAAUAAUUUUUGUUUUAUCUGUAACGCGUGCAU ((((.((((((((.((((.(((...........------)))))))...))).))))).))))....((((.(((...((((.((((........)))).)))).))).)))). ( -34.10) >DroYak_CAF1 15119 114 - 1 GUUGGCUGGUGUUAGGGAAUGUGUUUUGUGGGUUUGUUUCCAGCCCACCAGCAAUCAGGUGCCAACAUGCAGGUGAAAUGCAAAUAAUUUUUGUUUUAUCUGUAGCGCGGGUAU (((((.(((.(((.(((((((..((.....))..))))))))))))))))))......(((((..(.((((((((((..(((((.....)))))))))))))))..)..))))) ( -36.70) >consensus GGUGG___________GAAUGU_UU__GUGG_______UACAGCCCACCAGCAAUCAGGUGCCAACAUGCAGGUGAAAUGCAAAUAAUUUUUGUUUUAUCUGUAACGCGGGUAU ..........................................(((((((........)))((.....((((((((((..(((((.....)))))))))))))))..)))))).. (-19.20 = -19.68 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:24 2006