
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,754,198 – 4,754,296 |
| Length | 98 |
| Max. P | 0.998705 |

| Location | 4,754,198 – 4,754,296 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.22 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4754198 98 + 23771897 UUUGCCGGCACAUGCUGUUGCUGCAG----AACUGCAGCUGCGACAAAUAUUUAUUCAUAUUCAUGGGUAACAAACACAAUUUUUGUUUAUUUAUCGCGCCU------------------ ......(((..........((((((.----...)))))).((((.(((((..(((((((....)))))))(((((.......))))).))))).))))))).------------------ ( -27.00) >DroSec_CAF1 17623 97 + 1 UUUGCCGGCACAUGCUGUUGCUGCA-----AACUGCAGCUGCGACAAAUAUUUAUUCAUAUUCAUGGGUAACAAACACAAUUUUUGUUUAUUUAUCGCGCCU------------------ ......(((..........(((((.-----....))))).((((.(((((..(((((((....)))))))(((((.......))))).))))).))))))).------------------ ( -25.50) >DroSim_CAF1 15004 97 + 1 UUUGCCGGCACAUGCUGUUGCUGCA-----AACUGCAACUGCGACAAAUAUUUAUUCAUAUUCAUGGGUAACAAACACAAUUUUUGUUUAUUUAUCGCGCCU------------------ ......(((.......(((((....-----....))))).((((.(((((..(((((((....)))))))(((((.......))))).))))).))))))).------------------ ( -23.30) >DroEre_CAF1 6895 97 + 1 UUUGCCGGCACAUGCUGUUGCUGCA-----AACUGCAGCUGCUACAAAUAUUUAUUCAUAUUCAUGGGUAACAAACACAAUUUUUGUUUAUUUAUCGCGCCU------------------ ......(((.(.((..((.(((((.-----....))))).))..)).....((((((((....)))))))).(((((.......))))).......).))).------------------ ( -21.20) >DroYak_CAF1 13306 97 + 1 UUUGCCGGCACAUGCUGUUGCUGCA-----AACUGCAGCUGCGACAAAUAUUUAUUCAUAUUCAUGGGUAACAAACACAAUUUUUGUUUAUUUAUCGCGCCU------------------ ......(((..........(((((.-----....))))).((((.(((((..(((((((....)))))))(((((.......))))).))))).))))))).------------------ ( -25.50) >DroAna_CAF1 7208 119 + 1 UUUGCCGCCACAUGCUGUUGCUGCAACGCAAACUGCAGCUGCGACAAAUAUUUAUUCAUAUUCAUGG-AAGCAAAUACAAUUUUUGUUUAUUUAUCGCGCCUCACUGGGAUGGGGCUGUG ...((.(((.(((.((...((((((........)))))).((((.(((((....(((((....))))-)((((((.......))))))))))).))))........)).))).))).)). ( -31.60) >consensus UUUGCCGGCACAUGCUGUUGCUGCA_____AACUGCAGCUGCGACAAAUAUUUAUUCAUAUUCAUGGGUAACAAACACAAUUUUUGUUUAUUUAUCGCGCCU__________________ ......(((..........((((((........)))))).((((.(((((..(((((((....)))))))(((((.......))))).))))).)))))))................... (-22.22 = -22.92 + 0.70)



| Location | 4,754,198 – 4,754,296 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.22 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4754198 98 - 23771897 ------------------AGGCGCGAUAAAUAAACAAAAAUUGUGUUUGUUACCCAUGAAUAUGAAUAAAUAUUUGUCGCAGCUGCAGUU----CUGCAGCAACAGCAUGUGCCGGCAAA ------------------.(((((((((((((........(..((((..(.....)..))))..).....)))))))))).((((((...----.))))))..........)))...... ( -30.22) >DroSec_CAF1 17623 97 - 1 ------------------AGGCGCGAUAAAUAAACAAAAAUUGUGUUUGUUACCCAUGAAUAUGAAUAAAUAUUUGUCGCAGCUGCAGUU-----UGCAGCAACAGCAUGUGCCGGCAAA ------------------.(((((((((((((........(..((((..(.....)..))))..).....)))))))))).(((((....-----.)))))..........)))...... ( -28.72) >DroSim_CAF1 15004 97 - 1 ------------------AGGCGCGAUAAAUAAACAAAAAUUGUGUUUGUUACCCAUGAAUAUGAAUAAAUAUUUGUCGCAGUUGCAGUU-----UGCAGCAACAGCAUGUGCCGGCAAA ------------------.(((((((((((((........(..((((..(.....)..))))..).....))))))))((.(((((....-----....))))).))..)))))...... ( -26.52) >DroEre_CAF1 6895 97 - 1 ------------------AGGCGCGAUAAAUAAACAAAAAUUGUGUUUGUUACCCAUGAAUAUGAAUAAAUAUUUGUAGCAGCUGCAGUU-----UGCAGCAACAGCAUGUGCCGGCAAA ------------------.((((((...((((((((.......))))))))....(..(((((......)))))..).((.(((((....-----.)))))....)).))))))...... ( -26.40) >DroYak_CAF1 13306 97 - 1 ------------------AGGCGCGAUAAAUAAACAAAAAUUGUGUUUGUUACCCAUGAAUAUGAAUAAAUAUUUGUCGCAGCUGCAGUU-----UGCAGCAACAGCAUGUGCCGGCAAA ------------------.(((((((((((((........(..((((..(.....)..))))..).....)))))))))).(((((....-----.)))))..........)))...... ( -28.72) >DroAna_CAF1 7208 119 - 1 CACAGCCCCAUCCCAGUGAGGCGCGAUAAAUAAACAAAAAUUGUAUUUGCUU-CCAUGAAUAUGAAUAAAUAUUUGUCGCAGCUGCAGUUUGCGUUGCAGCAACAGCAUGUGGCGGCAAA ....(((((((....(((..(.((((((((((........(..(((((....-....)))))..).....)))))))))).(((((((......)))))))..)..))))))).)))... ( -34.72) >consensus __________________AGGCGCGAUAAAUAAACAAAAAUUGUGUUUGUUACCCAUGAAUAUGAAUAAAUAUUUGUCGCAGCUGCAGUU_____UGCAGCAACAGCAUGUGCCGGCAAA ...................(((((((((((((........(..(((((((.....)))))))..).....)))))))))).((((((........))))))..........)))...... (-26.30 = -26.52 + 0.22)

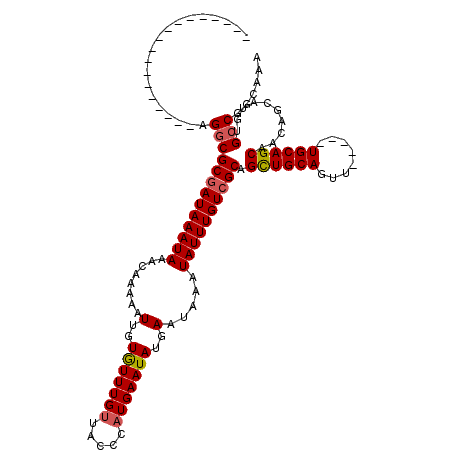
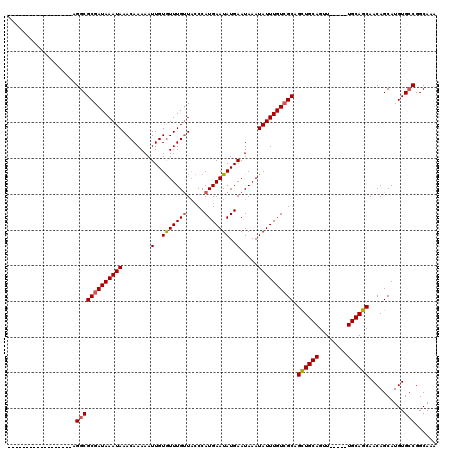
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:21 2006