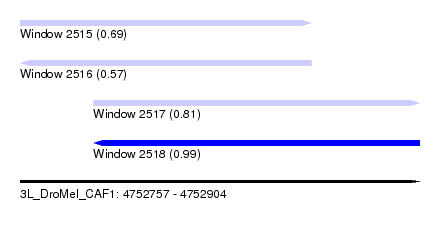
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 4,752,757 – 4,752,904 |
| Length | 147 |
| Max. P | 0.992359 |

| Location | 4,752,757 – 4,752,864 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -24.23 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4752757 107 + 23771897 GUGGAAAAAUCCAUC------------AGCA-AAAUAUUUGGCAUAAUAAUAAGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUGGUCAAUGGCCAAUGGCCAAGGGUCUGUUGCAAUUC (((((....))))).------------.(((-(.......(((.........(((..(((((...)))))..))).((((((.((((.....)))).))))))...)))..))))..... ( -32.70) >DroSec_CAF1 16133 118 + 1 GUGGAAAAAUCCAUCACUGGGUGGC-UAGCU-AAAUAUUUGGCAUAAUAAUAGGAGUGCAUGAUGCAUGCUGUCUAUGGCCAUUGGUCAAUGGCCAAUGGCCAAGGGUCUGUUGCAAUUC (..(....((((.....((((..((-..(((-((....)))))............((((.....))))))..))))(((((((((((.....))))))))))).))))...)..)..... ( -42.40) >DroSim_CAF1 13518 118 + 1 GUGGAAAAAUCCAUCACUGGGUGGC-UAGCA-AAAUAUUUGGCAUAAUAAUAGGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUGGUCAAUGGCCAAUGACCAAGGGUCUGUUGCAAUUC .(((....(((((....)))))..)-))(((-(....((((((((....(((((...(((((...)))))..)))))(((((........))))).))).)))))......))))..... ( -32.90) >DroEre_CAF1 5442 106 + 1 GUGGACAAGUCCAUCACUGGGGGAUAUAGAAAAAAUAUUUGGCAUAAUAAUAAGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUG--------------GCCAAGGGUCUGAUGCAAUUC (((((....))))).......((((((.......)))))).((((.......(((..(((((...)))))..))).(((((...)--------------))))........))))..... ( -23.00) >consensus GUGGAAAAAUCCAUCACUGGGUGGC_UAGCA_AAAUAUUUGGCAUAAUAAUAAGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUGGUCAAUGGCCAAUGGCCAAGGGUCUGUUGCAAUUC (((((....)))))...........................(((........(((..(((((...)))))..)))..((((..((((((........))))))..))))...)))..... (-24.23 = -24.47 + 0.25)

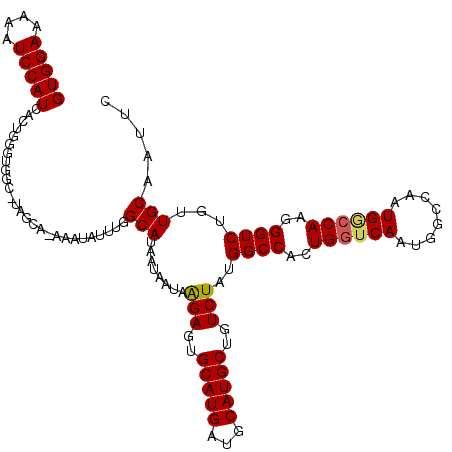
| Location | 4,752,757 – 4,752,864 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -23.10 |
| Energy contribution | -24.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4752757 107 - 23771897 GAAUUGCAACAGACCCUUGGCCAUUGGCCAUUGACCAGUGGCCAUAGACAGCAUGCAUCAUGCACUCUUAUUAUUAUGCCAAAUAUUU-UGCU------------GAUGGAUUUUUCCAC .....((((.......(((((...((((((((....)))))))).(((..(((((...)))))..))).........))))).....)-))).------------..((((....)))). ( -29.10) >DroSec_CAF1 16133 118 - 1 GAAUUGCAACAGACCCUUGGCCAUUGGCCAUUGACCAAUGGCCAUAGACAGCAUGCAUCAUGCACUCCUAUUAUUAUGCCAAAUAUUU-AGCUA-GCCACCCAGUGAUGGAUUUUUCCAC .....(((.......((((((((((((.......)))))))))).))...(((((...))))).............))).........-.....-.....(((....))).......... ( -28.40) >DroSim_CAF1 13518 118 - 1 GAAUUGCAACAGACCCUUGGUCAUUGGCCAUUGACCAGUGGCCAUAGACAGCAUGCAUCAUGCACUCCUAUUAUUAUGCCAAAUAUUU-UGCUA-GCCACCCAGUGAUGGAUUUUUCCAC .....((((..........(((..((((((((....))))))))..))).(((((...)))))........................)-)))..-.....(((....))).......... ( -28.10) >DroEre_CAF1 5442 106 - 1 GAAUUGCAUCAGACCCUUGGC--------------CAGUGGCCAUAGACAGCAUGCAUCAUGCACUCUUAUUAUUAUGCCAAAUAUUUUUUCUAUAUCCCCCAGUGAUGGACUUGUCCAC .................((((--------------(...)))))..(((((((((...))))).....................................(((....)))...))))... ( -19.00) >consensus GAAUUGCAACAGACCCUUGGCCAUUGGCCAUUGACCAGUGGCCAUAGACAGCAUGCAUCAUGCACUCCUAUUAUUAUGCCAAAUAUUU_UGCUA_GCCACCCAGUGAUGGAUUUUUCCAC .....(((.......((((((((((((.......)))))))))).))...(((((...))))).............)))............................((((....)))). (-23.10 = -24.10 + 1.00)

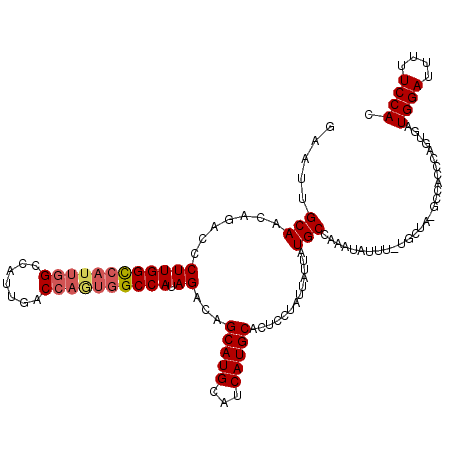
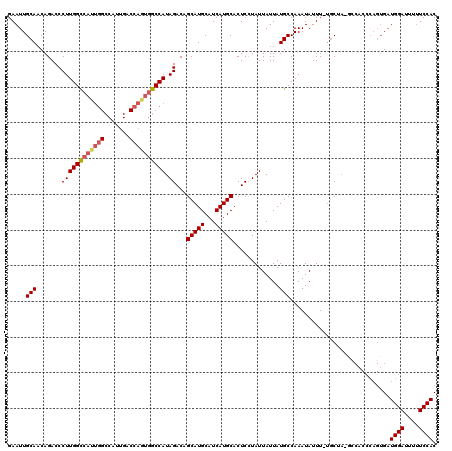
| Location | 4,752,784 – 4,752,904 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -35.89 |
| Consensus MFE | -30.20 |
| Energy contribution | -31.41 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4752784 120 + 23771897 GGCAUAAUAAUAAGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUGGUCAAUGGCCAAUGGCCAAGGGUCUGUUGCAAUUCUCCGAUUACCAAUGAUCCUUCCUCGGAUGGAUUGCUGAUU (((.........(((..(((((...)))))..))).((((((.((((.....)))).))))))...)))....((((((((((((.................))))).)))))))..... ( -39.43) >DroSec_CAF1 16171 120 + 1 GGCAUAAUAAUAGGAGUGCAUGAUGCAUGCUGUCUAUGGCCAUUGGUCAAUGGCCAAUGGCCAAGGGUCUGUUGCAAUUCUCCGAUUACCAAUUGUCCUUCCUCGGAUGGAUUGCUGAUU (((......(((((...(((((...)))))..)))))((((((((((.....))))))))))....)))....((((((((((((.................))))).)))))))..... ( -43.93) >DroSim_CAF1 13556 120 + 1 GGCAUAAUAAUAGGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUGGUCAAUGGCCAAUGACCAAGGGUCUGUUGCAAUUCUCCGAUUACCAAUGGUCCUUCCUCGGAUGGAUUGCUGAUU ((((.....(((((...(((((...)))))..)))))((((..((((((........))))))..))))))))((((((((((((..(((...)))......))))).)))))))..... ( -38.00) >DroEre_CAF1 5482 106 + 1 GGCAUAAUAAUAAGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUG--------------GCCAAGGGUCUGAUGCAAUUCUCCGAUUACCAAUGGUCCUGCCUCGGAUGGAUUGCUGAUU (((.........(((..(((((...)))))..))).(((((...)--------------))))...)))....((((((((((((........((.....))))))).)))))))..... ( -30.80) >DroYak_CAF1 11881 99 + 1 GGCAUAAUAAUAAGAGUGCAUGAUGCAUGCUGUCUAUGG---------------------CCAAGGGUCUGAUGCAAUUCUCCGAUUACCAAUGGUCCGUCCUCGGAUGGAUUGCUGAUU (((...(((...((.((((.....)))).))...))).)---------------------))...((((....((((((((((((..((.........))..))))).))))))).)))) ( -27.30) >consensus GGCAUAAUAAUAAGAGUGCAUGAUGCAUGCUGUCUAUGGCCACUGGUCAAUGGCCAAUGGCCAAGGGUCUGUUGCAAUUCUCCGAUUACCAAUGGUCCUUCCUCGGAUGGAUUGCUGAUU (((.........(((..(((((...)))))..))).((((((.((((.....)))).))))))...)))....((((((((((((.................))))).)))))))..... (-30.20 = -31.41 + 1.21)

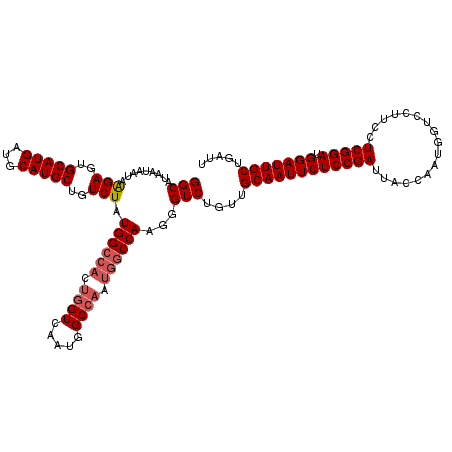
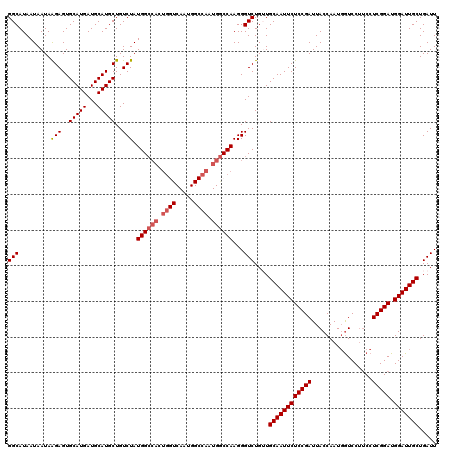
| Location | 4,752,784 – 4,752,904 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -30.12 |
| Energy contribution | -31.28 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 4752784 120 - 23771897 AAUCAGCAAUCCAUCCGAGGAAGGAUCAUUGGUAAUCGGAGAAUUGCAACAGACCCUUGGCCAUUGGCCAUUGACCAGUGGCCAUAGACAGCAUGCAUCAUGCACUCUUAUUAUUAUGCC .....(((((((.((....)).))))...(((((((.((((..............((((((((((((.......)))))))))).))...(((((...))))).)))).)))))))))). ( -38.50) >DroSec_CAF1 16171 120 - 1 AAUCAGCAAUCCAUCCGAGGAAGGACAAUUGGUAAUCGGAGAAUUGCAACAGACCCUUGGCCAUUGGCCAUUGACCAAUGGCCAUAGACAGCAUGCAUCAUGCACUCCUAUUAUUAUGCC .....(((.(((.((....)).)))....(((((((.((((..............((((((((((((.......)))))))))).))...(((((...))))).)))).)))))))))). ( -39.60) >DroSim_CAF1 13556 120 - 1 AAUCAGCAAUCCAUCCGAGGAAGGACCAUUGGUAAUCGGAGAAUUGCAACAGACCCUUGGUCAUUGGCCAUUGACCAGUGGCCAUAGACAGCAUGCAUCAUGCACUCCUAUUAUUAUGCC .....(((.(((.((....)).)))....(((((((.((((...(((....((((...))))..((((((((....))))))))......)))(((.....))))))).)))))))))). ( -38.60) >DroEre_CAF1 5482 106 - 1 AAUCAGCAAUCCAUCCGAGGCAGGACCAUUGGUAAUCGGAGAAUUGCAUCAGACCCUUGGC--------------CAGUGGCCAUAGACAGCAUGCAUCAUGCACUCUUAUUAUUAUGCC ..................((((((.((((((((....((.((......))...))....))--------------))))))))..(((..(((((...)))))..)))........)))) ( -26.10) >DroYak_CAF1 11881 99 - 1 AAUCAGCAAUCCAUCCGAGGACGGACCAUUGGUAAUCGGAGAAUUGCAUCAGACCCUUGG---------------------CCAUAGACAGCAUGCAUCAUGCACUCUUAUUAUUAUGCC .....(((((...(((((..((.((...)).))..)))))..)))))...........((---------------------(...(((..(((((...)))))..))).........))) ( -21.30) >consensus AAUCAGCAAUCCAUCCGAGGAAGGACCAUUGGUAAUCGGAGAAUUGCAACAGACCCUUGGCCAUUGGCCAUUGACCAGUGGCCAUAGACAGCAUGCAUCAUGCACUCUUAUUAUUAUGCC .....(((.(((.((....)).)))....(((((((.((((..............((((((((((((.......)))))))))).))...(((((...))))).)))).)))))))))). (-30.12 = -31.28 + 1.16)


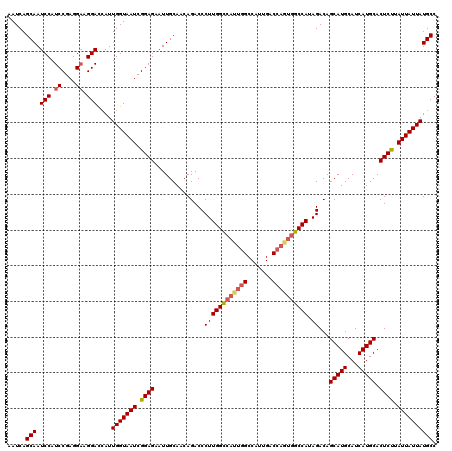
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:17 2006